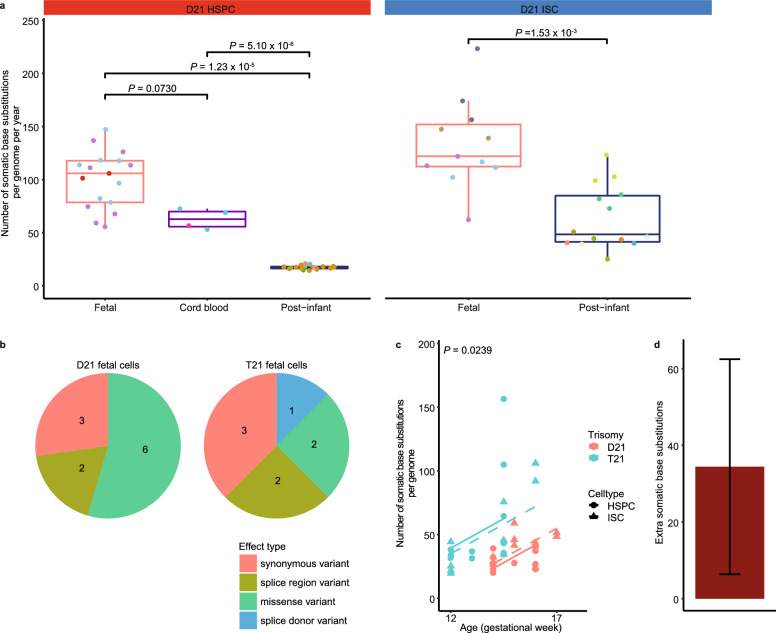
Figure 2.
Accumulation of somatic base pair substitutions in haematopoietic stem and progenitor cells (HSPCs) and intestinal stem cells (ISCs) during human fetal development and after birth. (a) Comparison of the number of autosomal somatic base substitutions per genome per year between D21 HSPCs (D21 fetal: 17 clones; 3 donors, Cord blood: 4 clones; 2 donors, Post-infant: 18 clones; 6 donors) and D21 ISCs (D21 fetal: 11 clones; 4 donors, Post-infant: 14 clones; 9 donors) of fetuses, cord blood and post infant (linear mixed-effects model). Points with the same color indicate single cells from the same subject. (b) Pie charts showing the number of somatic mutations for different types of exonic mutations in D21 and T21 fetal stem and progenitor cells (D21 fetal: 28 clones; 5 donors, T21 fetal: 23 clones; 4 donors). c The number of somatic base substitutions per genome plotted against the donor age (D21 fetal: 28 clones; 5 donors, T21 fetal: 23 clones; 4 donors). Dashed line: ISC, full line: HSPC. P-value shows the difference between T21 and D21 fetal stem and progenitor cells. (linear mixed-effects model, two-tailed t-test). d Extra somatic base substitutions per genome in T21 fetal stem and progenitor cells. Error bars represent 95% confidence intervals.