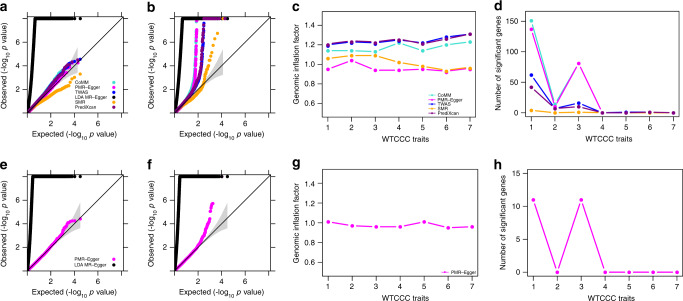
Fig. 4. TWAS analysis results by different methods for WTCCC traits.
Compared methods include CoMM (turquoise), PMR-Egger (magenta), TWAS (blue), LDA MR-Egger (black), SMR (orange), and PrediXcan (purple). a Quantile–quantile plot of −log10 p-values from different methods for testing the causal effect for an exemplary trait BD. b Quantile–quantile plot of −log10 p-values from different methods for testing the causal effect for another exemplary trait T1D. c Genomic inflation factor for testing the causal effect for each of the seven traits by different methods. d Number of causal genes identified for each of the seven traits by different methods. e Quantile–quantile plot of −log10 p-values from different methods for testing the horizontal pleiotropic effect for an exemplary trait BD. f Quantile–quantile plot of −log10 p-values from different methods for testing the horizontal pleiotropic effect for another exemplary trait T1D. g Genomic inflation factor for testing the horizontal pleiotropic effect for each of the seven traits by different methods. h Number of genes identified to have significant horizontal pleiotropic effect for each of the 7 traits by different methods. For c, d, g, h, the number on the x axis represents seven traits in order: T1D, CD, RA, BD, T2D, CAD, HT.