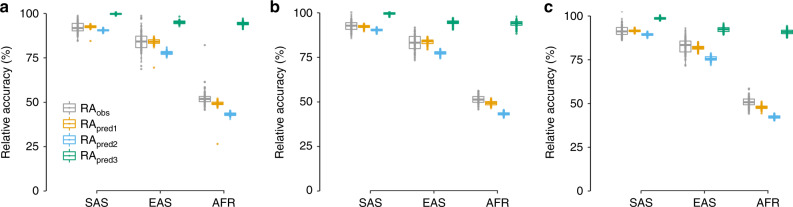
Fig. 2. Impact of negative selection on PGS trans-ancestry relative accuracies.
Relative accuracies (RA) of PGS in different ancestries under various strengths of negative selection. Traits were simulated with a heritability h2 = 0.5 and assuming MC = 5000 causal variants. Negative selection was modelled using a parameter S such that smaller values of S indicate stronger strength of selection. Values of S are denoted S1 and S2 in the discovery population and target populations, respectively. We considered thee scenarios: a S1 = S2 = −0.5; b S1 = −0.5, S2 = −0.75; and c S1 = −0.75, S2 = −0.5. RAobs, RApred1, RApred2 and RApred3 labels are defined as in the legend of Fig. 1. Boxes represent the first and third quantiles and whiskers are 1.5-folds the interquartile range. The points represent the RA for 100 replicates. The median estimates are shown as the horizontal line in the boxes.