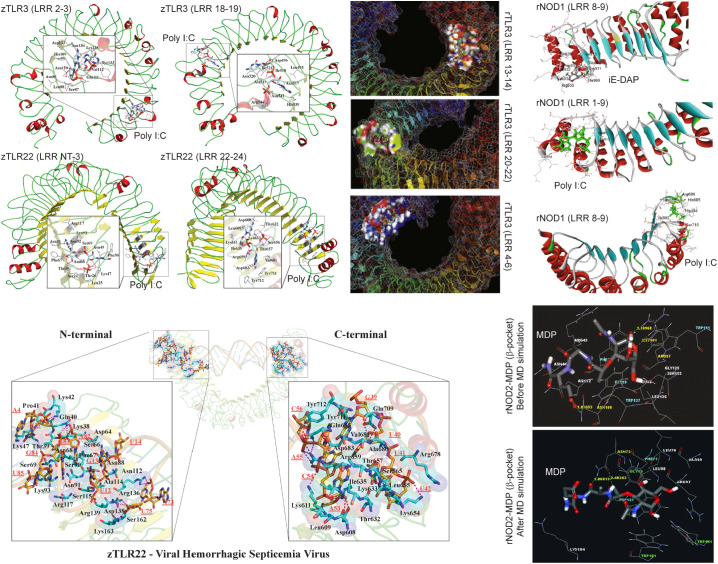
Fig. 6.
Structural interactions between different PAMPs and TLR and NOD receptors. Molecular docking simulations showing the binding sites of poly I:C in rohu and zebrafish TLR3-ECD and TLR22-ECD (referred to as rTLR and zTLR) predicted using AutoDock or GOLD programs. Molecular interaction of iE-DAP and poly I:C with rohu NOD1 (rNOD) predicted by molecular docking using the GOLD program. HADDOCK docking simulation illustrating the binding sites for the VHSV-dsRNA in the zebrafish TLR22-ECD. A comparative illustration of the MDP binding to rohu NOD2-LRR β-sheet pocket predicted from docking simulations (GOLD program) before and after MD simulations as indicated. The figures are reproduced from Ref. 44, 45, 102, and 103.