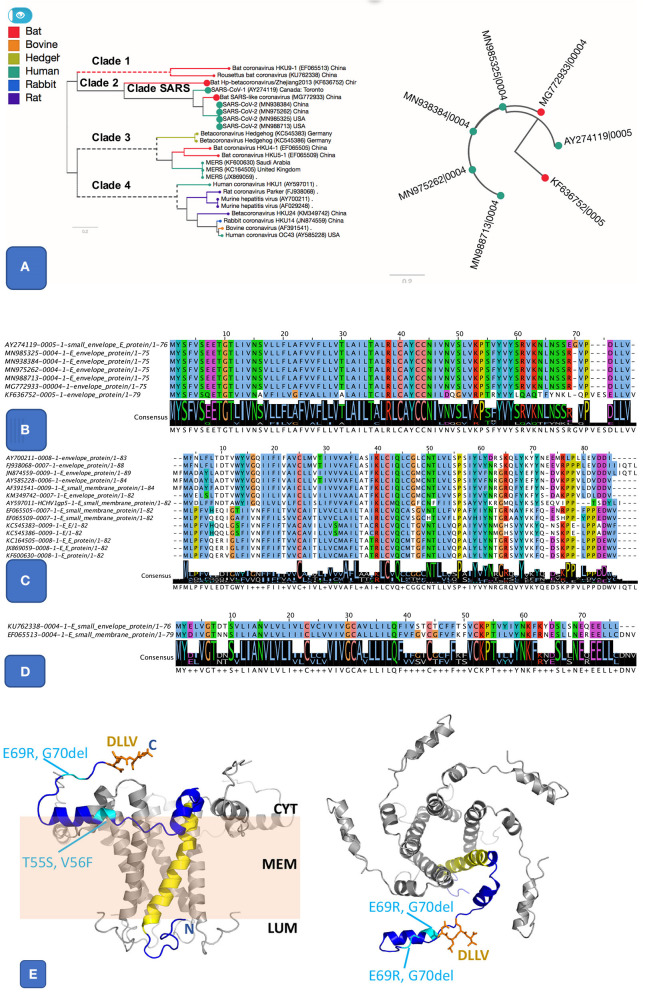
Figure 2.
Pangenome analysis of 3 clusters related E protein. (A) The first E cluster, it shows much similar E proteins from SARS and SARS-like genomes, highlighted in the species tree (A left panel) alongside a gene tree (A right panel). (B). Protein alignment of SARS and SARS-like E protein cluster. It includes SARS (AY274119), SARS-COV-2 (MN985325 and other isolates), and two bat coronaviruses (MG772933 and KF636752). The two important features are the ion-channel forming amino acids (where N15 and V25 were shown to be key for function) and the PBM class II motif (DLLV). Both features are completely conserved in SARS and SARS-CoV-2. (C) Protein alignment from another E cluster that groups E sequences from clade 3 and 4, including MERS and coronaviruses from other animals. (D) Protein alignment of 3rd E cluster that groups sequences from clade 1, related to two bat coronaviruses. (E) Theoretical model for the SARS-nCoV-1 protein E pentamer. Left Panel: side view. The membrane is illustrated in pale orange, and membrane (MEM), luminal side (LUM), and cytoplasmic side (CYT) are labeled. Right panel: view from the cytoplasm. One chain (blue) highlights protein features. N, C: location of N and C terminus, respectively. Yellow: IC; orange: DLLV (side chains are shown as stick models). Location of the residues changed in SARS-CoV-2 E are labeled and shown in cyan. The protein structure was modeled based on the NMR model PDB id 5 × 29, and completed using our in-house modeling program (to be published). The position of the region C-terminal to K53 was adjusted compared to the NMR model (see Supplementary Figure S6) to avoid its positioning entirely within the membrane, which appears unlikely given its amino acid composition (in particular R61, K63, and N64).