Figure 4. Increased stability of switch 1 may prevent mtEFG1 inhibition by fusidic acid.
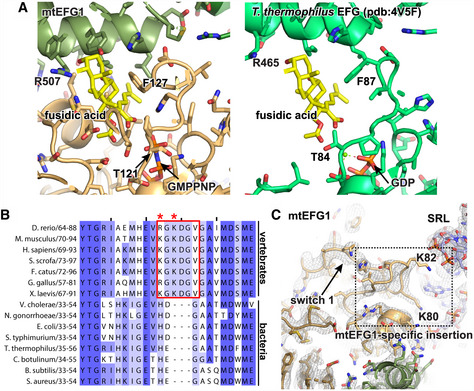
- Fusidic acid (yellow) has been modeled into its predicted binding site in mtEFG1 by superimposing a T. thermophilus model (pdb: 4V5F Gao et al, 2009). Critical residues for FA function are indicated. T121 and G127 correspond to T84 and F87 in T. thermophilus. The ordered conformation of switch 1 likely prevents binding of FA to mtEFG1. The bacterial structure, in which FA binding only occurs when switch 1 is disordered, is shown for comparison on the right (pdb:4V5F (Gao et al, 2009).
- Clustal Omega sequence alignment of the switch 1 region that contains the mtEFG1‐specific insertion from different vertebrate and bacterial species. Coloring was done according to percent identity in Jalview. The insert and K80 and K82 are highlighted with a red box or red asterisks, respectively.
- Switch 1 in the POST state is shown with the corresponding EM density at σ = 4. The position of the mtEFG1‐specific insertion with conserved K80 and K82 is boxed.
