Figure 6. Model for mtEFG1‐catalyzed mRNA‐tRNA translocation in mammalian mitochondria.
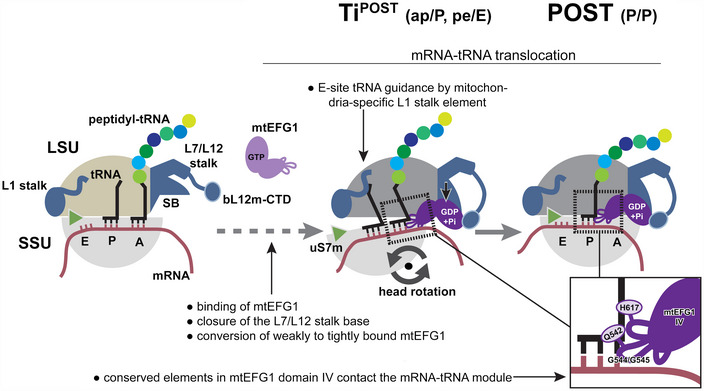
Following peptide bond formation, mtEFG1 is likely recruited to the ribosome by interaction with the bL12m‐CTD to catalyze translocation of the mRNA‐tRNA module. mtEFG1 binding to the ribosome induces a closure of the L7/L12 stalk base (SB), which converts mtEFG1 from a weakly to a tightly bound state that is translocation competent. Here, closure of the SB may be required to prolong the lifetime of the active GDP‐Pi state of mtEFG1. mRNA‐tRNA movement depends on large‐scale motions of the ribosome. Swiveling of the head repositions the tRNAs on the SSU ‐ the deacylated P site tRNA moves into the chimeric pe position and the peptidyl‐tRNA in the A site into the ap position, respectively. Translocation is completed upon backrotation and backswiveling of the SSU body and head, respectively, which positions the peptidyl‐tRNA into the classical P/P conformation. mtEFG1 contains highly conserved elements at the tip of domain IV that are required for interaction with the tRNA backbone and the minor groove of the mRNA‐tRNA module. Domain IV maintains these contacts throughout the translocation process. The dashed arrow indicates that in analogy to the bacterial system likely multiple additional translocation intermediates exist preceding the ones visualized in this study.
