Figure EV1. Prediction and validation of OTU DUBs from bacteria.
-
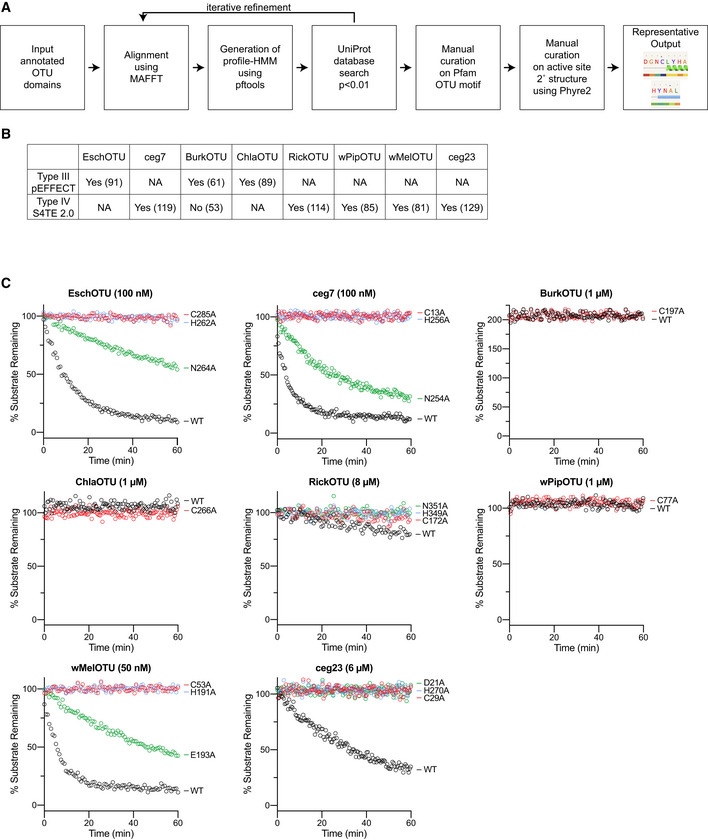
AWorkflow illustrating the process used for bioinformatic prediction of bacterial OTU domains, followed by their manual curation. Representative output from the Phyre2 curation is shown for the predicted EschOTU active site.
-
BTable presenting prediction scores for type III and type IV secretion signals using the pEFFECT (Goldberg et al, 2016) and S4TE 2.0 (Noroy et al, 2019) prediction approaches, respectively. For pEFFECT predictions, prediction reliability scores above 50 or above 80 are associated with 87% or 96% accuracy, respectively. For S4TE 2.0, a prediction score above 72 is associated with a 98% sensitivity. Organisms that lack one of the secretion systems are marked as not applicable (NA).
-
CFull fluorescent Ub substrate cleavage data for all bacterial OTUs following Ala substitution at each member of the predicted catalytic triad. These data were collected in parallel with those presented in Fig 1G, and the WT dataset is shown again for clarity. The rise above 100% observed with BurkOTU is indicative of a noncovalent interaction.
