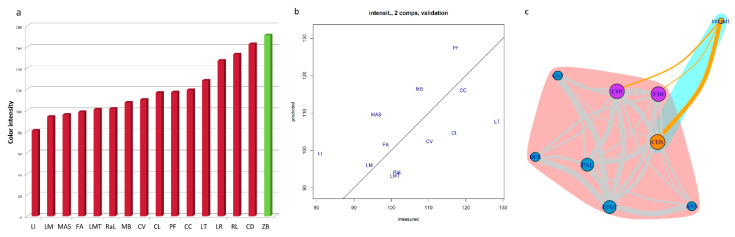
Figure 6.
Statistical analyses of the quantitative trait of anthocyanins accumulation in petals. The accessions are indicated with codes: ‘Incomparabile’—LI, ‘Meyer’—LM, M. australasica var. sanguineaMAS, ‘Femminello Adamo’—FA, ‘Mexican lime’—LMT, ‘Rangpur lime’—RaL, ‘Buddha’s hand’—MB, C. volkameriana—CV, Citrus latipes—CL, ‘Pink fleshed’—PF, C. celebica—CC, ‘Tahiti’—LT, ‘Lima rossa corrugata’—LR, ‘Red lime’—RL, ‘Diamante’—CD, ‘Zagara bianca’—ZB. (a) Bar plot of petal color intensity measured by image analysis on petals as the intensity in grayscale (whitest has the highest value) reported for each accession. (b) Validation plot of the partial least square regression (PLS) model for petal pigmentations. Values measured by image analysis of petals versus predicted values from gene expression using the first two components of the model. (c) The network of gene expressions in petals involved in pigmentation measured as the color intensity. The thickness of edges represents the Pearson’s Correlation Coefficient value, the node dimension represents the weight of the node in the network, the color of the edge represents the relationship among genes in grey and with the phenotypical trait in dark orange. The node colors represent the subgroups and the halo represents community detection, based on edge-betweenness.