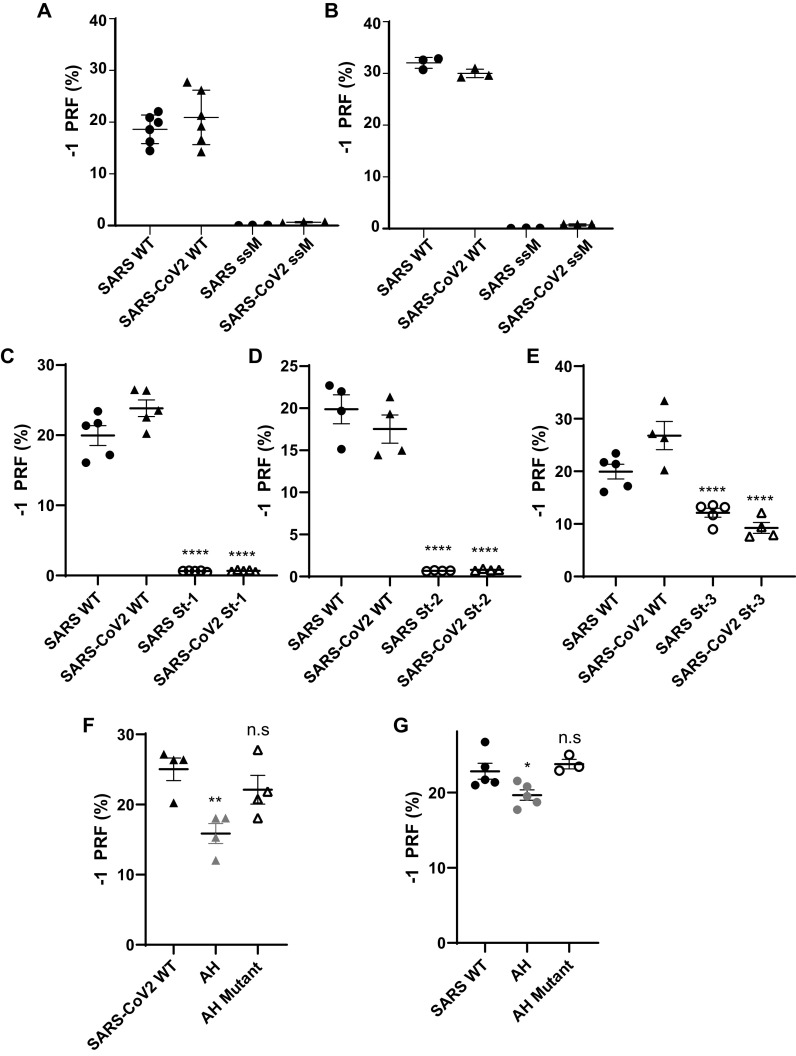
Figure 2.
Functional characterization of the SARS-CoV and SARS-CoV-2 −1 PRF signals. A and B, analyses of silent slippery site mutants. The efficiencies of −1 PRF promoted by the WT (U UUA AAC) and silent slippery site mutant (C CUC AAC) −1 PRF signals were assayed in HEK (A) and HeLA (B). ssM denotes silent slippery site mutant. C–E, analyses of the importance of the three stems in the −1 PRF stimulating RNA pseudoknot. Silent stem 1 (St-1, C), stem 2 (St-2, D), and stem 3 (St-3, E) mutants were assayed in HEK cells. F and G, analyses of the attenuator hairpins. AH denotes constructs that included attenuator hairpin sequences. AH mutant denotes mutants harboring the silent coding attenuator hairpin sequences shown in Fig. 1 (E and F). Assays were performed using Dual-Luciferase assays as previously described (15, 16). Each data point represents a single biological replicate comprised of three technical replicates. Error bars denote S.E. n.s, not significant.