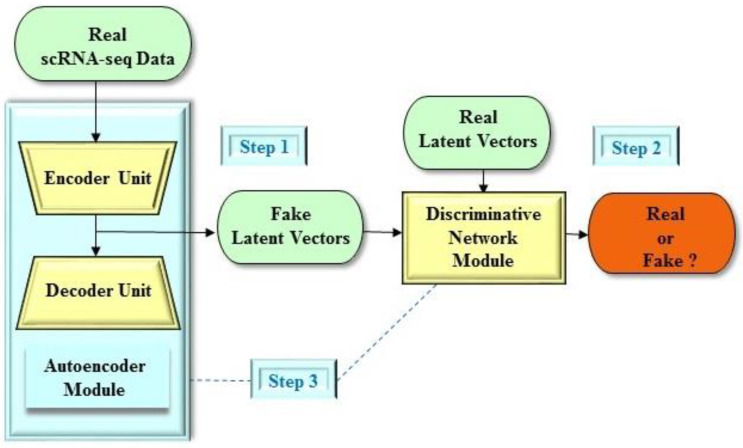
Figure 5.
An example of the deep adversarial variational autoencoder structure for dimensionality reduction in scRNA-seq analysis. Step 1: The encoder unit produces synthetic latent vectors as real as possible. The encoder unit provides the mean and covariance of the Gaussian distribution to serve as the variational distribution, which is commonly generated by a variational autoencoder structure. Step 2: The discriminative network module assesses the probability that the latent vector stems from the real latent vectors. Step 3: Both the autoencoder and discriminative network modules play concurrently against each other to obtain their objectives.