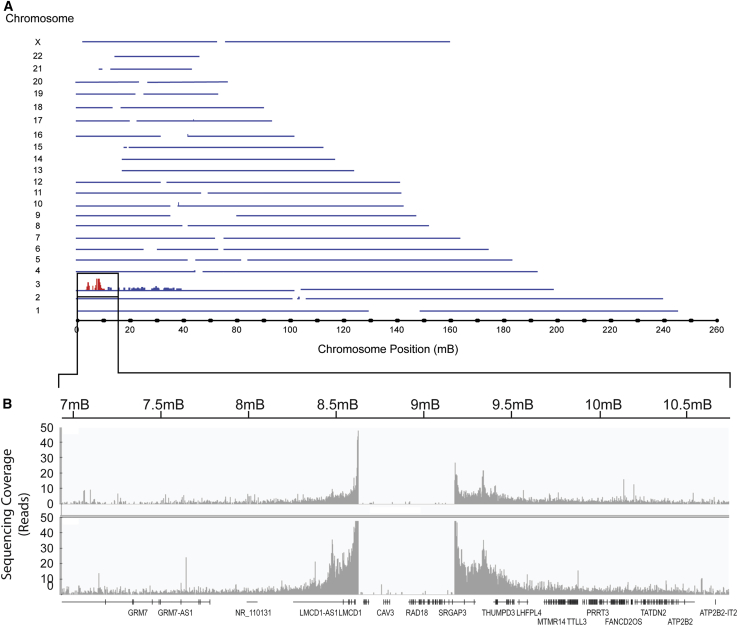
Figure 4.
Analysis of Knockout Clones by Targeted Sequencing
(A) Targeted sequencing of T antigen integration sites. The plot shows TLA sequence coverage across the HEK293T cell clone D9 genome using primers targeting the pRTAK plasmid origin of replication. The single plasmid integration site present on chromosome 3 (chr3) of the HEK293T D9 cell clone is shown in red. (B) TLA sequence coverage of the plasmid integration site referred to in (A). The x axis shows genomic features from human chr3: 6,938,850–10,764,483. The two boxplots with gray bars indicate sequence coverage observed when enrichment was conducted with primers targeting the origin of replication (upper boxplot) or T antigen-encoding sequences (lower boxplot). The y axis is limited to 50-fold coverage. Data in this figure are from the parental D9 cell clone, but they are representative of deletion clones #109 and #126, as they yielded similar integration sites. Box magnified area is not to scale.