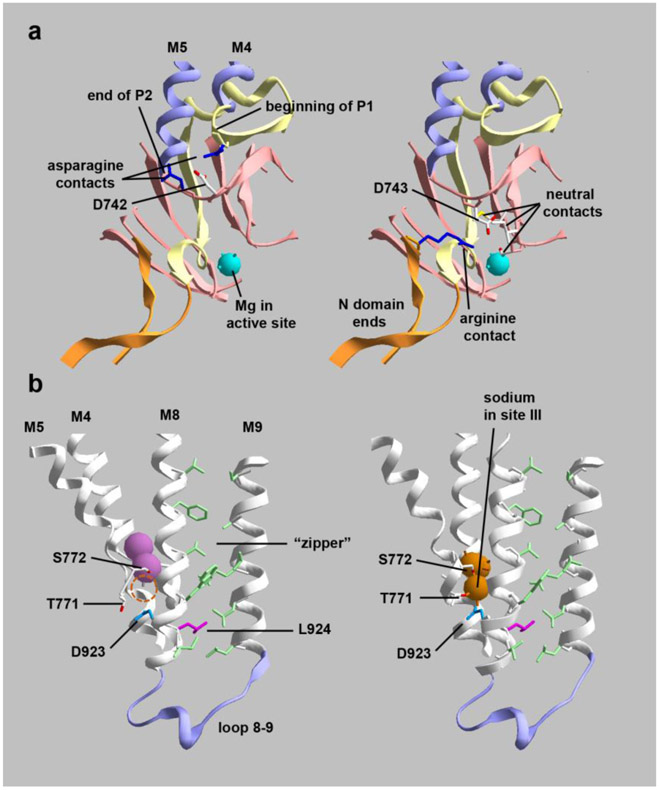
Figure 2. Cutaway images of crystal structures showing the positions and interactions of mutated residues.
(a) D742 and D743 are adjacent aspartates at a junction between protein domains. They are on an unstructured link between the second part of the P domain (P2) and transmembrane span M5. (left) D742 (colored in CPK) has polar interactions with two asparagines (dark blue), one at the beginning of P1 and one at the end of P2. Substitution with a bulky tyrosine (D742Y) may impede the folding of the two parts of the P domain. (right) D743 (also colored in CPK) points down toward the active site where Mg+2 is positioned. D743 interacts with 3 local residues in P2 and with an arginine (dark blue) located at the beginning of the N domain; D743 and R586 do not appear to form a strong salt bridge. Substitution with hydrophilic histidine may not affect protein folding but may impact the active site. (b) D923N (blue) and L924P (magenta) are adjacent residues that also point in different directions. (left) K+-bound conformation (2 pink spheres); (right) Na+-bound conformation (3 orange spheres). D923 on M8 is close to T771 and S772 on M5, residues that bind the third sodium ion. The empty sodium binding site is marked with a dashed circle in the K+ conformation. T771 and S772 produce AHC when mutated, while D923N produces RDP or AHC. D923N is well-known to have residual activity and to reduce affinity for sodium (Einholm et al., 2010; Holm et al., 2016). L924 (magenta) has a distinct structural role. While most transmembrane spans in polytopic membrane proteins intersect each other at angles, M8 and M9 are very parallel. They are held together by a zipper-like structure of opposing residues (light green). L924 (magenta) interacts with two opposing leucines in the zipper near its base where the M8-M9 hairpin establishes the position of the 8-9 loop, an important part of the structure. Substitution of L924 with a proline will disrupt the helix at that position, so it is likely to cause misfolding and may displace both D923 and the 8-9 loop. The structures are PDB codes 2ZXE (Shinoda et al., 2009) and 3WGU (Kanai et al., 2013). [color essential]