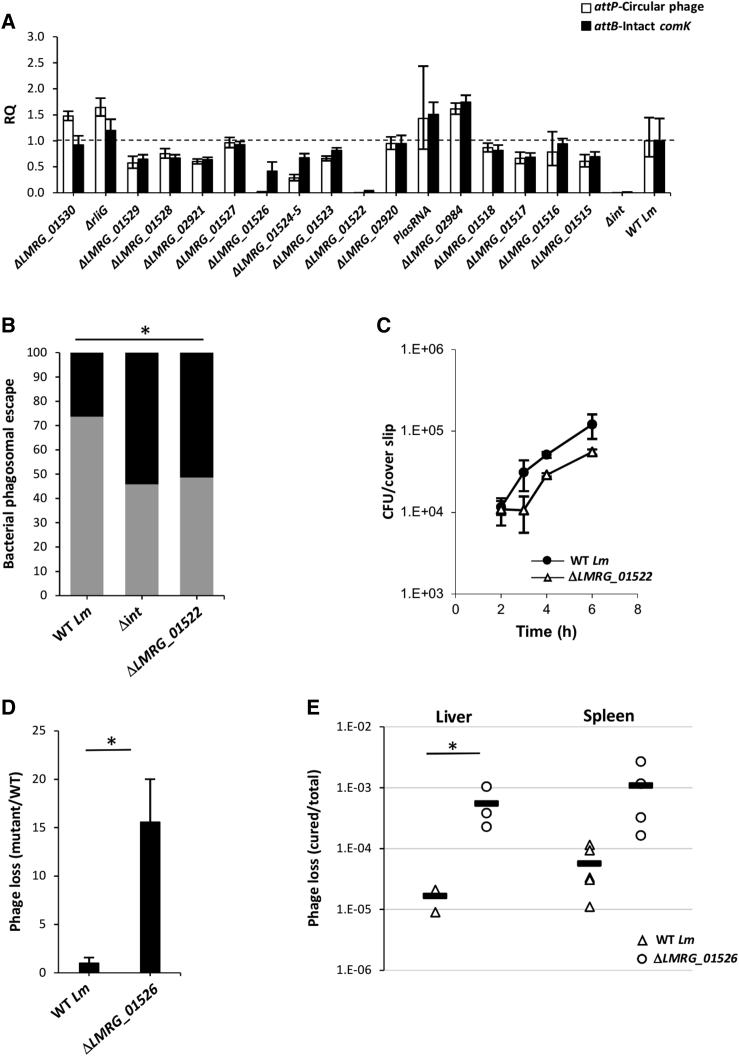
Figure 4.
Phage Early Genes That Contribute to Active Lysogeny
(A) qRT-PCR analysis of attB and attP sites in WT Lm and indicated mutants grown in BMDM cells for 6 h. Data are presented as relative quantity (RQ), compared to the levels in WT bacteria. The data represent a mean of three independent experiments. Error bars indicate the 95% confidence interval.
(B) A bacterial phagosomal escape assay. Percentage of bacteria that escaped the macrophage phagosomes as determined by a microscope fluorescence assay. Macrophages were infected with WT Lm and ΔLMRG_01522, as well as with Δint as a control. The data represent two biological repeats. ∗p < 0.05 as calculated by the χ-test.
(C) Intracellular growth of WT Lm and ΔLMRG_01522 mutant in BMDM cells. The data are representative of three independent experiments. Error bars represent the standard deviation of a triplicate.
(D) Analysis of phage loss in ΔLMRG_01526 mutant in comparison to WT Lm grown in macrophage cells for 6 h. The number of phage-cured bacteria isolated from macrophages infected with ΔLMRG_01526 mutant was normalized to that of WT bacteria. The data represent the mean of three independent experiments. Error bars represent the standard deviation. ∗p < 0.05 as calculated by Student’s t test.
(E) Analysis of phage loss in the ΔLMRG_01526 mutant in comparison to WT Lm during in vivo infection of C57BL/6 mice (48 h.p.i.). The number of phage-cured bacteria isolated from the spleens and livers of infected mice was normalized to the total number of bacteria recovered. The data represent 3 to 5 mice per sample. ∗p = 0.05 as calculated by Student’s t test.