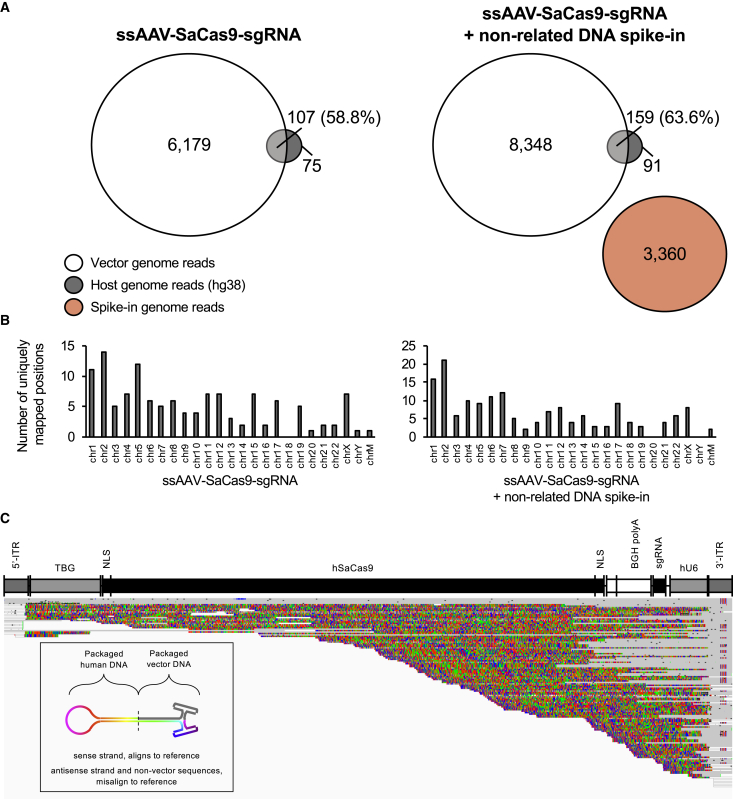
Figure 3.
Evaluation of Host-Cell DNAs Encapsidated within ssAAV-SaCas9-sgRNA Vectors
(A) Venn diagrams of mapped read abundances related to the vector genome (white circles), the host-cell genome (gray circles), or a non-related DNA spike-in (red circle). Non-overlapping portions represent reads that map exclusively to either reference. Regions of overlap represent the counts of reads that co-mapped to both the host-cell genome and the vector genome. The percentages of co-mapped reads are displayed. (B) Histograms summarizing the number of unique regions throughout the host-cell genome (hg38) to which sequencing reads are mapped. Left graphs, without non-related DNA spike-in; right graphs, with spike-in. (C) Squished IGV display of chimeric reads mapped to the vector genome. Sequence regions that align to the vector reference are in gray, while those that do not are colored as their respective bases. A unifying feature of chimeric reads is that they are anchored at the 3′ ITR region. The lower left schematic illustrates the hypothesized self-complementary chimeric structures for reference.