Figure 6. Identification of the Subunits of PP2A Required for the Phosphatase Activation and Antitumor Activities of iHAP1 and SMAP.
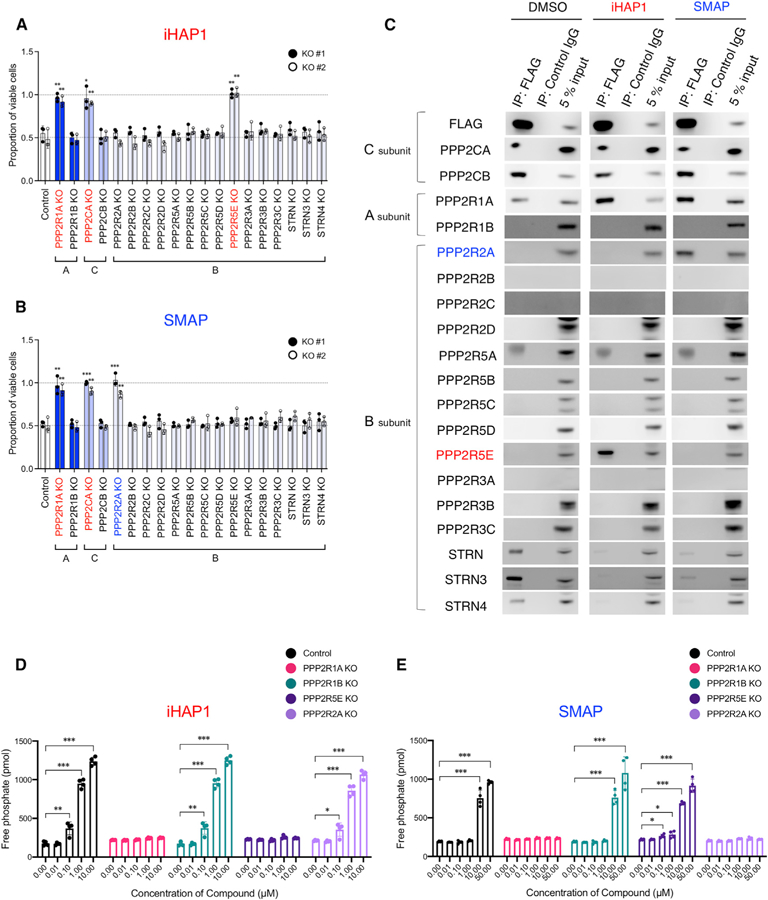
(A and B) Sensitivity to (A) iHAP1 and (B) SMAP, as tested in KOPT-K1 cells with selective inactivation of PP2A subunits. Each subunit was knocked out by CRISPR-Cas9 with use of two unique gRNAs designed for each subunit (#1 and #2; Figure S6). Control gRNA targeted the luciferase gene. Cells were treated with 0.5 µM iHAP1 or 5 µM SMAP for 72 h and then examined for viability. *p < 0.05, **p < 0.01, and ***p < 0.001 by Student’s t test, comparing the means ± SD of three biological replicates versus controls.
(C) Coimmunoprecipitation assays using an anti-FLAG antibody to pull down N-terminal FLAG-tagged PPP2CA and PPP2CB expressed in KOPT-K1 cells. Cell lysates were treated with 1 µM iHAP1, 10 µM SMAP, or DMSO for 1 h at room temperature. Then PP2A subunits were coimmunoprecipitated with the FLAG-tagged C subunits and detected by western blotting with antibodies specific for each subunit.
(D and E) Phosphatase activity of PP2A in control KOPT-K1 cells compared with KOPT-K1 cells with selective PP2A subunit inactivation. Cells were treated with (D) iHAP1 or (E) SMAP at various concentrations for 3 h. KO indicates knockout of the gene in KOPT-K1 cells. Values are shown as means ± SD of four biological replicates versus controls. *p < 0.05, **p < 0.01, and ***p < 0.001 by Student’s t test.
