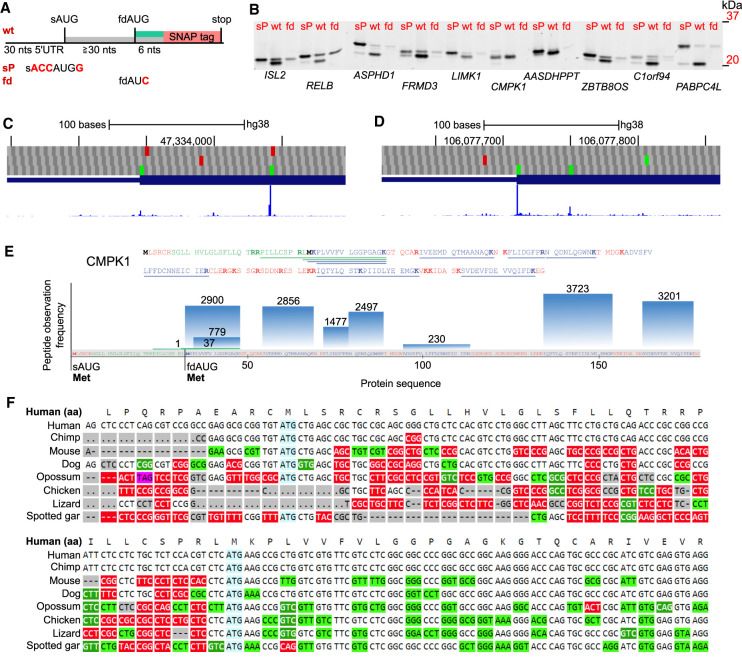
Figure 4.
Experimental verification of downstream initiation at mRNAs containing sAUGs in evolutionarily conserved weak context. (A) Schematic of the test sequence cassette fused to SNAP tag. The wt test sequence includes 30 nt upstream of sAUG, the spacer between sAUG and fdAUG, and 6 nt downstream from fdAUG. For each wt test sequence there are two controls, sP has the sAUG in perfect Kozak context, and fd has the fdAUG changed to AUC. wt, sP, and fd test cassettes were designed for 10 selected genes. (B) Scans of protein gels used to separate SNAP-tagged protein products from test constructs expressed in HEK293T cells. Gene names are shown below the lanes. (C,D) GWIPS-viz screenshots for CMPK1 and AASDHPPT loci, respectively. The top plots show codons in three reading frames with AUGs colored green and stop codons colored red. Below is RefSeq annotation of corresponding transcripts, and the thicker area of bars represents CDS. GWIPS-viz aggregated initiating ribosomes track is at the bottom, showing the density of footprints obtained from the ribosomes enriched at initiating sites with specific drugs. (E) Peptide observation frequency from PeptideAtlas for CMPK1 protein. In red are tryptic peptides not expected to be detectable. In green are detectable peptides unique for the long proteoform, and in blue are shared peptides. (F) Codon alignment of representative mammalian genomic sequences in the vicinity of sAUG and fdAUG (highlighted in blue) of CMPK1 locus. Synonymous substitutions are highlighted in light green; nonsynonymous are shown in white font highlighted in green for similar and in red for dissimilar amino acids according to BLOSUM62.