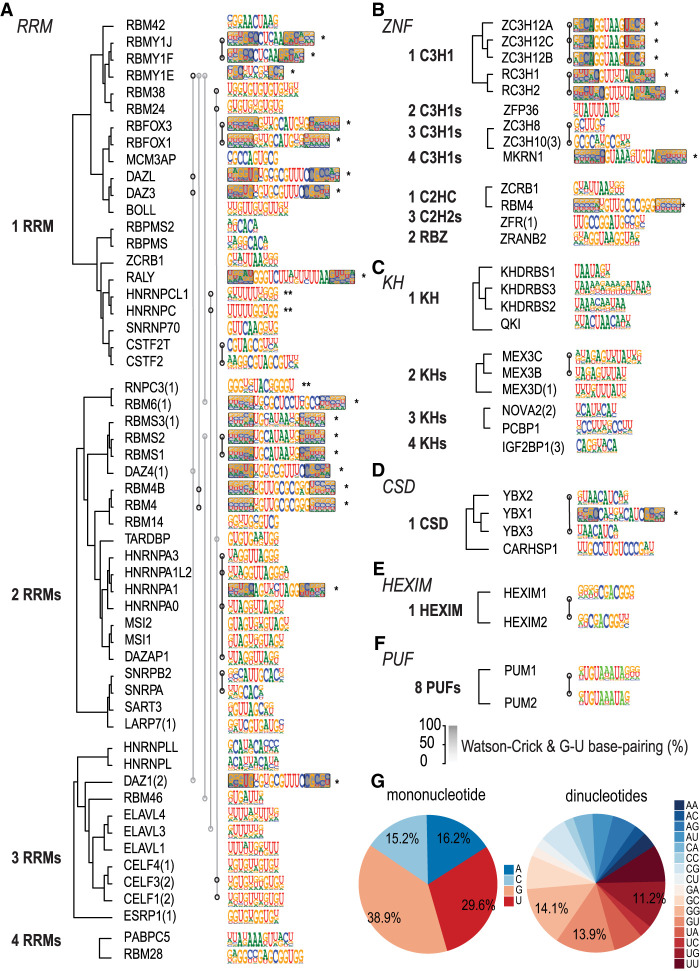
Figure 4.
Comparison between the HTR-SELEX motifs. (A–F) Similar RBPs bind to similar motifs. Motifs were classified into six major categories based on structural class of the RBPs. Dendrograms are based on amino acid alignment using PRANK (Löytynoja and Goldman 2005). Within the RRM family, RBPs with different numbers of RRMs were grouped and aligned separately; if fewer RBDs were included in the construct used, the number of RBDs is indicated in parentheses (see also Supplemental Table S1). Motifs shown are the primary motif for each RBP. Asterisks indicate a stem–loop structured motif, with the gray shading showing the strength of the base-pairing at the corresponding position. Two asterisks indicate that the RBP can bind to a structured secondary motif. Motifs that are similar to each other based on SSTAT analysis (covariance threshold 5 × 10−6) are indicated by open circles connected by lines. Only families with more than one representative HTR-SELEX motif are shown. (G) RBPs commonly prefer sequences with G or U nucleotides. Frequencies of all mononucleotides (left) and dinucleotides (right) across all of the RBP motifs. Note that G and U are overrepresented.