Table 1.
Structure of viral proteins which were used as a targets for molecular docking with PDB ID (4-character unique identifier of every entry in the Protein Data Bank) and source organism.
| Name of the protein and description | Source | PDB ID | Structure of protein |
|---|---|---|---|
| Mpro/3CLpro (Main protease of SARS CoV 2) | SARS CoV 2 | 6Y84 | 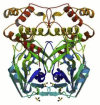 |
| The crystal structure of COVID-19 main protease | SARS CoV 2 | 6LU7 | 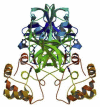 |
| The crystal structure of COVID-19 main protease in apo form | SARS CoV 2 | 6M03 |  |
| Nsp10 (Plays a crucial role in viral transcription by stimulating both nsp14 3′-5′ exoribonuclease and nsp16 2′-O-methyltransferase activities.) Nsp 16 (S-adenosylmethionine-dependent (nucleoside-2′-O)-methyltransferase only active in the presence of its activating partner nsp10) |
SARS CoV 2 | 6W75 B 6W75D 6W75 A 6W75D |
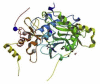 |
| Spike glycoprotein | SARS CoV 2 | 6VSBA 6VSBB 6VSBC |
 |
| SARS protein receptor binding domain | SARS CoV | 2GHV | 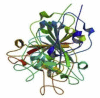 |
| Nsp9 (RNA binding protein of SARS CoV 2) and SARS CoV 2 helicase | SARS CoV 2 | 6W4B |  |
| ADP ribose phosphatase of NSP3 from SARS CoV-2 | SARS CoV 2 | 6VXS |  |
| Angiotensin-converting enzyme 2 (ACE2) receptor (Receptor through which virus enters the cell) | Human | 6M18B 6M18D |
 |
