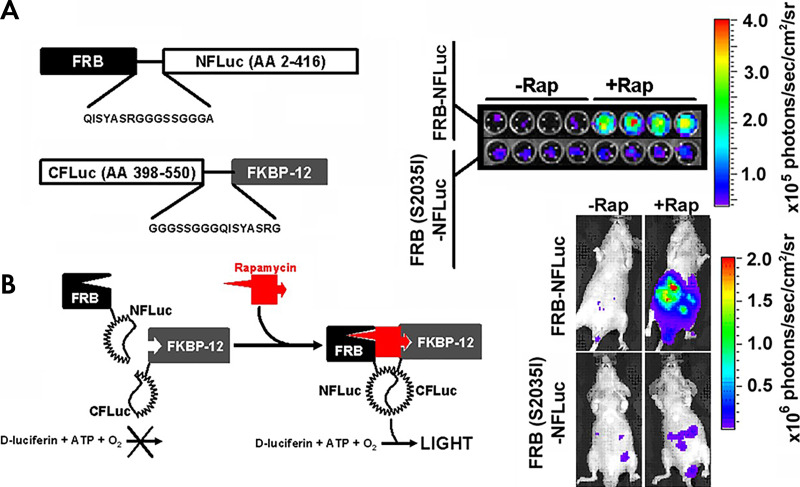
Figure 3:
Luciferase complementation imaging. Genetically encoded luciferase complementation strategies enable noninvasive imaging of reversible protein-protein interactions and protein folding events in cellulo and in vivo (44,106). Multiple protein-protein interactions can be monitored through the use of multicolor click beetle luciferases by combining complementation strategies and spectral unmixing (45). A, The two interacting proteins are fused to the N-terminal fragment of the luciferase and the C-terminal fragment of the luciferase, respectively, with an interposed flexible glycine-serine linker. In this example, the two interacting proteins are the rapamycin-binding protein (FKBP) and FKBP rapamycin binding domain (FRB) that associate in the presence of rapamycin. When they associate, the luciferase active site is reconstituted, and light is produced. B, Rapamycin-induced light production is specific both in cellulo (top) and in vivo (bottom). A mutation known to abrogate the binding of rapamycin (S2035I) inhibits light production both in cellulo and in vivo. (Reprinted, with permission, from reference 7.) ATP = adenosine triphosphate, CFLuc = C domain of the luciferase, NFLuc = N domain of the luciferase, Rap = rapamycin.