Abstract
Craniofacial anomalies are among the most common birth defects and are associated with increased mortality and, in many cases, the need for lifelong treatment. Over the past few decades, dramatic advances in the surgical and medical care of these patients have led to marked improvements in patient outcomes. However, none of the treatments currently in clinical use address the underlying molecular causes of these disorders. Fortunately, the field of craniofacial developmental biology provides a strong foundation for improved diagnosis and for therapies that target the genetic causes of birth defects. In this chapter, we discuss recent advances in our understanding of the embryology of craniofacial conditions, and we focus on the use of animal models to guide rational therapies anchored in genetics and biochemistry.
1. MODELS TO UNCOVER GENETICS OF CLEFT LIP AND PALATE
Cleft lip with or without cleft palate (CL/P) is the most common congenital anomaly of craniofacial development, affecting approximately 1 in 700 live births (Dixon, Marazita, Beaty, & Murray, 2011; Iwata, Parada, & Chai, 2011; Jugessur, Farlie, & Kilpatrick, 2009). It is estimated that 70% of CL/P cases are isolated and nonsyndromic and that 30% occur as part of one of more than 500 Mendelian syndromes (Dixon et al., 2011). Over the past few decades, mouse models have proved an invaluable tool in understanding the etiology of CL/P (Table 1; Bush & Jiang, 2012; Gritli-Linde, 2008; Jiang, Bush, & Lidral, 2006). These models have led to exponential growth in our knowledge of the molecular and cellular basis for clefts and have contributed greatly to the understanding of general developmental mechanisms. To date, the principle clinical benefit of information gained from these studies has been to improve diagnostics and estimates of recurrence risks (Dixon et al., 2011; Grosen et al., 2010). Looking toward the future, our improved understanding of the developmental processes underlying cleft pathogenesis increasingly suggests opportunities for developing new preventative and therapeutic approaches.
Table 1.
Clefting Genes and Candidate Loci with Associated Mouse Models of Orofacial Clefting
| Human Disease | Gene/Locus | Mouse Phenotype |
|---|---|---|
| Nonsyndromic CL/P | MSX1 | Cleft palate (Satokata & Maas, 1994) |
| Nonsyndromic CL/P | TGFβ3 | Cleft palate (Kaartinen et al., 1995; Proetzel et al., 1995) |
| Nonsyndromic CL/P | IRF6 | CP (Ingraham et al., 2006; Richardson et al., 2006) |
| Nonsyndromic CL/P | 8q24 locus | Deletion of 8q24 region in mice results in CL/P possibly by loss of Myc regulation (Uslu et al., 2014) |
| Nonsyndromic CL/P | PDGFC | CP (Ding et al., 2004) |
| Nonsyndromic CL/P | VAX1 | CP (Bertuzzi, Hindges, Mui, O’Leary, & Lemke, 1999) |
| Nonsyndromic CL/P | ARHGAP29 (ABCA4 locus) | CP (Leslie et al., 2012) |
| Nonsyndromic CL/P | BMP4 | Conditional ablation in the epithelium results in CL (Liu et al., 2005) |
| Nonsyndromic CL/P | FGFR2 | Conditional ablation in the epithelium results in CP (Hosokawa et al., 2009; Rice et al., 2004) |
| Nonsyndromic CL/P | MYH9 | Conditional ablation in the epithelium results in fusion defects; compound homozygous loss of Myh9; Myh10 results in CP (Kim et al., 2015) |
| Syndromic CL/P | Gene/Locus | Mouse Phenotype |
| Van Der Woude syndrome (OMIM#119300) | IRF6 | CP (Ingraham et al., 2006; Richardson et al., 2006) |
| Ankyloblepharon–ectodermal defects–cleft lip/palate (AEC) (OMIM#106260); Ectrodactyly, ectodermal dysplasia, and cleft lip/palate(EEC) (OMIM#129900) | P63 | Knock-in of human allele results in autosomal dominant CP and phenocopies AEC (Ferone et al., 2012), and homozygous null displays CL/P and phenocopies EEC (Moretti et al., 2010; Thomason, Dixon, & Dixon, 2008) |
| Loeys–Dietz syndrome (OMIM#609192) | TGFBR1, TGFBR2, SMAD3, TGFB2 | Conditional ablation of TgfBR1 or TgfBR2 in palate epithelium or NCC results in CP (Dudas et al., 2006; Ito et al., 2003; Iwata et al., 2012; Xu et al., 2006) |
| Stickler type 1 (OMIM#108300) | COL2A1 | CP (Li, Prockop, et al., 1995) |
| Stickler type 2 (OMIM#604841) | COL11A1 | CP (Li, Lacerda, et al., 1995; Seegmiller, Fraser, & Sheldon, 1971) |
| Smith–Lemli–Opitz syndrome (OMIM#270400) | DHCR7 | CP phenocopies SLOS (Wassif et al., 2001) |
| Treacher Collins (OMIM#154500) | TCOF1 | CP phenocopies Treacher Collins (Dixon et al., 2006; Jones et al., 2008) |
| Craniofrontonasal syndrome (CFNS) (OMIM#304110) | EFNB1 | CP phenocopies CFNS (Bush & Soriano, 2010; Compagni, Logan, Klein, & Adams, 2003) |
| Pierre Robin syndrome (OMIM#261800) | SOX9 | CP (Bi et al., 2001) |
| Andersen Syndrome (OMIM#170390) | KCNJ2 | CP (Zaritsky, Eckman, Wellman, Nelson, & Schwarz, 2000) |
| Cleft palate with or without ankyloglossia, X-linked (CPX) (OMIM#303400) | TBX22 | CP phenocopies CPX (Pauws et al., 2009) |
| DiGeorge syndrome (OMIM#188400) | TBX1 | CP phenocopies DiGeorge (Jerome & Papaioannou, 2001) |
| Branchiooculofacial syndrome (OMIM#113620) | TFAP2a | Chimeras and hypomorphic embryos have CL/P (Green et al., 2015; Nottoli, Hagopian-Donaldson, Zhang, Perkins, & Williams, 1998), ablation from NCC results in CP (Brewer, Feng, Huang, Sullivan, & Williams, 2004), deletion from midface results in midline cleft (Nelson & Williams, 2004), hypomorphic allele results in CL/P |
| Greig cephalopolysyndactyly syndrome (OMIM#175700) | GLI3 | CP (Huang, Goudy, Ketova, Litingtung, & Chiang, 2008) |
| Holoprosencephaly 9 (OMIM#610829) | GLI2 | CP (Mo et al., 1997) |
| Coloboma, heart anomaly, choanal atresia, retardation, genital and ear anomalies (CHARGE) (OMIM#214800) | CHD7 | CP phenocopies CHARGE (Sperry et al., 2014) |
| Hypogonadotropic hypogonadism with or without anosmia (Kallman syndrome) (OMIM#147950) | FGFR1 | CP (Trokovic, Trokovic, Mai, & Partanen, 2003) or CP with midline cleft (Wang et al., 2013) |
| Crouzon syndrome(OMIM#101200) | FGFR2 | CP, phenocopies Crouzon (Eswarakumar, Horowitz, Locklin, Morriss-Kay, & Lonai, 2004) |
| Apert syndrome (OMIM#101200) | FGFR2 | CP, phenocopies Apert (Martinez-Abadias et al., 2013) |
| Otopalatodigital syndrome (OMIM#311300) | FLNA | CP (Hart et al., 2006) |
| Lymphedema-distichiasis syndrome (OMIM#153400) | FOXC2 | CP (Iida et al., 1997; Winnier, Hargett, & Hogan, 1997) |
| Hypothyroidism, athyroidal, with spiky hair and cleft Palate (OMIM#241850) | FOXE1 | CP, models hereditary thyroid dysgenesis and cleft palate (De Felice et al., 1998) |
| Glass syndrome (OMIM#612313) | SATB2 | CP phenocopies 2q32-q33 deletion syndrome (Britanova et al., 2006) |
| Saethre–Chotzen syndrome (OMIM#101400) | TWIST1 | Ablation in mandibular NCCs results in posterior CP (Zhang et al., 2012) |
Complete tables of human clefting genes and loci, including those without mouse models can be found in Dixon et al. (2011).
From an embryologic perspective, formation of the lip and palate has developmental similarities in that both require the coordinated growth and fusion of embryonic prominences. However, these are temporally distinct events that involve different morphogenetic programs. The development of the lip occurs during the 4th through 6th week of human development and involves fusions of the maxillary, medial nasal, and lateral nasal processes, whereas secondary palate development occurs between the 6th and 12th week and involves the fusion of the maxillary-derived secondary palatal shelves (Bush & Jiang, 2012; Jiang et al., 2006). Consistent with this temporal asynchrony, cleft lip and cleft palate can occur separately or together; cleft lip is defined as a gap between the philtrum and the lateral upper lip, and cleft palate is defined as a gap in the secondary palate (roof of the mouth). Although the lip and palate form through separate developmental programs, many genetic factors play a role in both processes.
Prior to the advent of genome-wide methodologies, candidate genes for linkage and mutation analysis were selected largely on the basis of phenotypes arising from gene-targeted mutations in mice. For example, targeted mutation of the Msx1 homeobox gene resulted in syndromic cleft secondary palate and tooth agenesis in homozygous mutant mice and led to the evaluation of its human ortholog as a human CL/P candidate (Satokata & Maas, 1994). Linkage disequilibrium was found between MSX1 and human CL/P, and ultimately, mutations in MSX1 were identified in patients with syndromic cleft palate with tooth agenesis as well as in patients with non-syndromic CL/P; in total, mutations in MSX1 may contribute to around 2% of nonsyndromic CL/P cases (Jezewski et al., 2003; Lidral et al., 1998). Integration of mouse genetics, ex vivo, and molecular techniques has advanced our understanding of the developmental basis for Msx1 involvement in CL/P. Msx1 controls cell proliferation in the anterior secondary palate by maintaining mesenchymal expression of Bmp4 in the palatal mesenchyme (Alappat, Zhang, & Chen, 2003; Parada & Chai, 2012; Zhang et al., 2002). Whereas mutations in MSX1 have been identified in humans with cleft lip, Msx1−/− mice do not exhibit cleft lip, but rather cleft palate only. The related MSX2 might partially compensate for MSX1 loss in mice, because mice lacking MSX1 and MSX2 exhibit bilateral CL/P as part of dramatic craniofacial dysmorphogenesis (Ishii et al., 2005). Interestingly, Msx1−/− and Pax9−/− mice exhibit cleft lip with variable penetrance, possibly due to a shared role of MSX1 and PAX9 in the regulation of Bmp4 or other factors critical for lip morphogenesis (Liu et al., 2005; Nakatomi et al., 2010; Zhou et al., 2013). On the other hand, compound loss of Dlx5−/− and Msx1−/− in mice restored cell proliferation and rescued the cleft palate phenotype (Han et al., 2009). These results exemplify the complexity of the genetic networks controlling lip and palate development and support the likely importance of genetic interaction in human CL/P. Indeed, PAX9 has been confirmed as a contributor to human syndromic and nonsyndromic CL/P (Ichikawa et al., 2006; Schuffenhauer et al., 1999). Mouse genetics studies designed to elucidate genetic pathways have therefore been highly useful in identifying candidate genes for human genetic analysis.
The TGFβ3 gene has been considered as a candidate human clefting gene based on the cleft palate phenotype in Tgfβ3−/− homozygous mice (Kaartinen et al., 1995; Proetzel et al., 1995). Though no loss-of-function mutations have been identified in TGFβ3 in nonsyndromic cleft patients, association with the TGFβ3 genomic locus has been demonstrated (Lidral et al., 1998). Interestingly, treatment of mouse Tgfβ3−/− homozygous palatal shelves with exogenous TGFβ3 in explant culture resulted in a rescue of palate fusion, even when treated with a low dose for a relatively short time (Taya, O’Kane, & Ferguson, 1999). Further, exogenous TGFβ3 induced fusion of explanted lip primordia in CL/Fr, a mouse line that exhibits high susceptibility to cleft lip with unknown genetic etiology (Juriloff & Fraser, 1980; Kohama, Nonaka, Hosokawa, Shum, & Ohishi, 2002; Muraoka et al., 2005). These studies suggest the possibility of modulation of TGFβ signaling as a potential therapeutic approach for nonsyndromic CL/P.
Notably, mutations in other TGFβ pathway components including TGFB2, SMAD3, TGFβR2, or TGFβR2 cause syndromic cleft palate as part of autosomal dominant Loeys–Dietz syndrome (LDS; Loeys et al., 2005). The majority of LDS cases are caused by mutations in TGFβR1 or TGFβR2 and result in impaired kinase activity without altered expression or localization of the receptor, though the net effect is an apparent increase in TGFβ signaling (Loeys et al., 2005; Van Laer, Dietz, & Loeys, 2014). Marfan syndrome (MFS) has significant clinical overlap with LDS, but does not include clefting phenotypes and is also thought to involve overactive TGFβ signaling (Dietz et al., 1991; Habashi et al., 2006). In mice, TGFβ-neutralizing antibodies or the angiotensin II type 1 receptor (AT1) blocker losartan was used to decrease TGFβ signaling, which led to successful treatment of aortic aneurism. On this basis, several trials have been conducted in MFS patients to determine whether suppressing TGFβ signaling can reduce aortic aneurism (Bowen & Connolly, 2014; Chiu et al., 2013; Groenink et al., 2013; Habashi et al., 2006; Lacro et al., 2014). It will be interesting to determine whether similar improvements can be obtained for craniofacial phenotypes in LDS mouse models. However, prenatal use of such a strategy for LDS is likely to be complicated by embryonic consequences of blockade of TGFβ signaling and by timing of diagnosis, which raises an important general point: many of the genes involved in clefting have pleiotropic roles in the embryo, and it is not trivial to devise treatments that would lead to blockade of the pathway in one organ but not others.
During secondary palate fusion in mice, signaling by TgFβR2 is required for the expression of the gene encoding the IRF6 transcription factor, and this regulation is required for normal palate development (Iwata et al., 2013). Human mutations in IRF6 cause Van der Woude syndrome (VWS), an autosomal dominant ectodermal dysplasia that causes CL/P with lip pits (Kondo et al., 2002). Linkage between IRF6 and nonsyndromic CL/P has been shown in a number of studies, supporting its importance in human clefting (Dixon et al., 2011; Zucchero et al., 2004). Mice harboring mutations orthologous to those causing VWS phenocopy the disease and exhibit defects in epithelial differentiation that result in oral adhesions and cleft palate (Ingraham et al., 2006; Richardson et al., 2006; Stottmann, Bjork, Doyle, & Beier, 2010). Interestingly, expression of Irf6 is regulated by the transcription factor P63 in mice, and mutations in P63 in humans cause ectrodactyly–ectodermal dysplasia clefting (EEC) and ankyloblepharon ectodermal dysplasia clefting (AEC). Further, mice harboring compound heterozygous mutations in Irf6 and p63 exhibit fully penetrant cleft palate, connecting these ectodermal dysplasias to VWS through a molecular mechanism.
More recently, several genome-wide association studies (GWAS) have identified a number of additional loci that are likely to be involved in non-syndromic cleft lip and palate (Beaty et al., 2010, 2011; Birnbaum et al., 2009; Grant et al., 2009; Mangold et al., 2010). In addition to confirmation of IRF6 as a key susceptibility locus, four previously unidentified loci, including candidate genes with previously unknown involvement in craniofacial development, were identified (Mangold, Ludwig, & Nothen, 2011). Interestingly, all of these studies identified a locus at 8q24 as having significant involvement in nonsyndromic CL/P. Subsequent work in mice has indicated that this region includes a remote cis-acting enhancer for the Myc transcription factor (Uslu et al., 2014). Deletion of this region resulted in a dramatic decrease in expression of Myc specifically in the developing lip and a low frequency of CL/P, suggesting that regulatory mutations in MYC may contribute to human clefting linked to the 8q24 locus (Uslu et al., 2014). Estimation of the contribution of the four loci identified in these studies (IRF6, 8q24, 17q22, and 10q25.3) suggests that these loci might account for approximately 55% of variation in CL/P (Marazita, 2012). These data may challenge the traditional view of CL/P as a highly multifactorial disorder, raising new hopes for designing therapeutics if mechanistic insights for these loci can be obtained.
Gene–environment interactions have long been known to be a significant contributor to orofacial clefting in humans, and maternal smoking and alcohol consumption, as well as nutritional deficiency, have been implicated (Dixon et al., 2011). The extent of this contribution is not clear, however, and few susceptibility loci have been identified consistently. Recently, a GWAS approach has been used to identify gene–environment interactions underlying nonsyndromic cleft palate. Several loci, which harbor genes with previously unrecognized roles in orofacial clefting, were shown to have significant interaction with maternal smoking or alcohol use, whereas these loci did not reach significance if considered independently of smoking or alcohol use (Beaty et al., 2011). Mouse teratogen studies have shown that environmental influences can contribute to orofacial clefting, but very few studies have brought modern molecular and cell biological approaches to bear on understanding the mechanisms of these interactions. Such studies will be critical for understanding the mechanisms of action for these genes and determining if preventative approaches can reduce the frequency of clefting in humans.
2. TREACHER COLLINS: PROOF OF CONCEPT OF A NONSURGICAL THERAPEUTIC FOR A CRANIOFACIAL SYNDROME
Treacher Collins syndrome (TCS) is a congenital disorder that affects craniofacial development. Mutations in treacle (TCOF1) were the first identified cause of TCS, and since then additional genetic causes have been identified as well (Dixon, Trainor, & Dixon, 2007). Over 120 mutations in TCOF1, half of which occur spontaneously, have been identified, and most result in a premature stop codon, suggesting that haploinsufficiency of TCOF1 underlies the pathogenesis of TCS (Dixon et al., 2007).
Individuals with TCS exhibit a spectrum of craniofacial anomalies, including hypoplasia of the facial bones, particularly the mandible and zygomatic complex; downward slanting of the palpebral fissures, usually with colobomas of the lower eyelids and a paucity of lashes medial to the defect; abnormalities of the external ears and atresia of external auditory canals, resulting in hearing loss; and choanal atresia. Cleft palate and absence of the zygomatic arch may occur in severe cases (Edwards et al., 1996; Fazen, Elmore, & Nadler, 1967; Phelps, Poswillo, & Lloyd, 1981; Rovin, Dachi, Borenstein, & Cotter, 1964). There is high phenotypic variability among TCS individuals, and some individuals are retrospectively diagnosed after their more severely affected siblings or children are born.
Tcof1 expression in mouse embryos coincides with the formation and migration of neural crest cells, suggesting a role in their development (Dixon et al., 2006). Studies of neural crest cells in Tcof1+/− embryos revealed that, although the migration pattern remained unchanged, there was a reduction in the number of migrating neural crest cells compared to wild-type embryos (Dixon et al., 2006). Consistent with this observation, there was a significant increase in apoptosis and reduction in proliferation of neuroepithelial and neural crest cells in Tcof1+/− embryos. Interestingly, the half-life and cytoplasmic concentration of P53 were increased in Tcof1+/− embryos, and proapoptotic genes that are transcriptional targets of P53 showed increased expression levels as well (Jones et al., 2008).
Genetically ablating one or both alleles of Trp53, which encodes P53, in Tcof1+/− embryos resulted in dosage-dependent rescue of the TCS-like phenotypes (Jones et al., 2008). All Tcof1+/−;Trp53−/− pups survived beyond weaning with a fully rescued craniofacial phenotype. However, some of the pups developed tumors, which are commonly observed in Trp53−/− mice (reviewed in Levine, 1997). Chemical inhibition of P53 with pifithrin-α at the onset of neural crest cell migration led to a significant decrease in neuroepithelial apoptosis and subsequent alleviation of TCS-related phenotypes, suggesting the exciting possibility that small molecules administered in utero could treat TCS (Jones et al., 2008).
The successful rescue experiments in the TCS mouse model provide a conceptual advance in terms of medical treatment of birth defects resulting from ribosome abnormalities. This work also suggests that inhibition of P53-mediated apoptosis is a potential strategy for prenatal treatment of TCS. However, the strong correlation between P53 inhibition and tumorigenesis indicates that targeting P53 itself is not likely to be a practical therapeutic. Instead, investigating the TCOF effectors that are more specifically involved in TCS pathogenesis would be the logical next step to develop a safe and effective therapeutic for TCS.
3. RASopathies: UNDERSTANDING AND DEVELOPING TREATMENT FOR SYNDROMES OF THE RAS PATHWAY
The RASopathies are a group of syndromes characterized by dysregulation of signaling through the RAS pathway and include neurofibromatosis type 1 (NF1), Noonan syndrome (NS), NS with multiple lentigines, capillary malformation–AV malformation syndrome, Legius syndrome, Costello syndrome (CS), and cardio-facio-cutaneous syndrome (CFC; Fig. 1; Tidyman & Rauen, 2009). The RASopathies are caused by mutations in genes that encode components of the RAS pathway, and these conditions have both unique and overlapping characteristics.
Figure 1.
Syndromes of the RAS pathway. Schematic of the RAS signaling pathway with dashed lines connecting the syndrome with the protein in the pathway encoded by the causative mutated gene. Syndromes described in the text. NS, Noonan syndrome; NF1, neurofibromatosis 1;CFC, cardio-facio-cutaneous syndrome;CS, Costello syndrome.
The RAS genes, HRAS, NRAS, and KRAS, encode small, monomeric GTPases that are activated by upstream regulators, including receptor tyrosine kinases, G-protein-coupled receptors, and integrins. Once activated, RAS-GTP signals through multiple effector pathways, including RAF/MEK/ERK, PI3K/AKT, TIAM1/Rac, and RALGDS/Ral. Mouse models have advanced our understanding of the in vivo role of the RAS signaling pathway as well as how it is regulated, and animal models have also pointed to therapeutic targets for the RASopathies. Here, we focus on three of the RASopathies, NS, CS, and CFC, all of which have important craniofacial phenotypes.
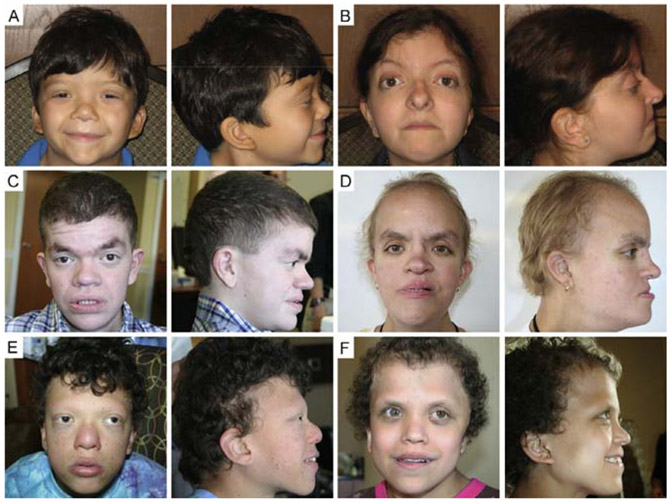
NS typically presents with proportional short stature, facial dysmorphia, including a typical “triangular facies” with dolicocephaly, prominent forehead, pointed chin and hypertelorism, as well as blood and cardiovascular abnormalities (Fig. 2A and B; Collins & Turner, 1973; Noonan, 1968; Zenker, 2009). In approximately 50% of NS cases, mutations in the PTPN11 gene, which encodes the SHP2 phosphatase, cause pathogenesis by increasing phosphatase activity (Tartaglia et al., 2001); other genes known to cause NS include SOS1 (~10%) (Roberts et al., 2007; Tartaglia et al., 2007), RAF1 (3–5%) (Pandit et al., 2007; Razzaque et al., 2007), KRAS (1–2%) (Schubbert et al., 2006; Zenker et al., 2007), NRAS (<1%) (Cirstea et al., 2010), and SHOC2 (<1%) (Cordeddu et al., 2009). Several mouse models with mutations in the genes responsible for NS have revealed the different contributions of specific genes to the syndrome pathology and shed light on the complexity of the RAS pathway. Mice expressing the NS-associated mutation Ptpn11D61G (Ptpn11D61G/+) have decreased body weight and length, craniofacial dysmorphia including decreased skull length, increased intercanthal distance and wide, blunt snout, myeloproliferative disease (MPD), and a range of cardiac defects (Araki et al., 2004). These mice do not develop hypertrophic cardiomyopathy (HCM), reflecting the fact that only 10% of NS individuals with PTPN11 mutations develop HCM. However, approximately 95% of patients carrying RAF1 mutations that cause increased kinase activity develop HCM, and knock-in mice expressing the kinase-activating NS mutation Raf1L613V have short stature, facial dysmorphia including short nose and wide-set eyes, and these mice develop HCM (Wu et al., 2011). Mice with the Sos1E846K-activating mutation display increased embryonic and perinatal mortality, short stature, craniofacial dysmorphia with short skull length, increased intercanthal distance with blunt snout due to depression of the frontal bone, and severe cardiac hypertrophy (Chen et al., 2010). Another strain of mice carrying an endogenous K-RasV14I germline mutation, the most frequent KRAS mutation in NS individuals, display many of the phenotypic abnormalities observed in NS, including small size, craniofacial dysmorphism with increased skull width and height but decreased length, wide separation between the eyes and blunt snout, and cardiac defects (Hernandez-Porras et al., 2014). Moreover, these mice develop fatal MPD, a disease similar to a leukemia seen in patients with NS. Although these NS mouse models share craniofacial characteristics similar to the triangular facies of NS individuals, the cardiac conditions vary depending upon the gene mutation, emphasizing the specific effects of distinct gene mutations.
Figure 2.
Craniofacial features of the RASopathies. The RASopathies are caused by mutations in genes encoding different proteins in the RAS pathway and have both unique and overlapping craniofacial characteristics. (A) Frontal and profile photographs of a 7-year-old male and (B) 21-year-old female with Noonan syndrome (NS), (C) a 20-year-old male and (D) 23-year-old female with Costello syndrome (CS), and (E) a 15-year-old male and (F) 15-year-old female with cardio-facio-cutaneous syndrome (CFC).
CS is characterized by craniofacial malformations including macrocephaly, bitemporal narrowing, convex facial profile, full cheeks and large mouth, dermatologic anomalies, cardiac defects, musculoskeletal abnormalities, growth delay, and cognitive deficits (Fig. 2C and D; Goodwin, Oberoi, et al., 2014; Rauen, 2007). Nearly all individuals with CS have a heterozygous, de novo germline mutation in HRAS that results in a constitutively active protein (Aoki et al., 2005; Estep, Tidyman, Teitell, Cotter, & Rauen, 2006). The majority of the mutations encode a HRASG12S protein. There are no mouse models expressing HRASG12S; however, mice expressing HRASG12V, the mutation most commonly expressed in HRAS tumors (Schubbert, Bollag, & Shannon, 2007), phenocopy and provide an effective model of CS. A mouse model with a germline G12V mutation within the endogenous H-Ras locus (HrasG12V/geo) had several characteristics of CS, including craniofacial dysmorphia due to depression of the anterior frontal bone, shortening and depression of the nasal bridge and premaxillary bone, and choanal atresia, and HCM including cardiac enlargement and hypertrophic cardiomyocytes (Schuhmacher et al., 2008). Another CS mouse model using a targeted conditional activating HrasG12V mutation (CC/FR-HrasG12V) displayed many traits associated with CS, including failure to thrive and decreased growth rate, relative acrocephaly, deviated septum, and myocardial fibrosis (Chen et al., 2009). These mice also exhibited an enamel defect characterized by hyperproliferative and disorganized ameloblasts similar to the hypoplastic enamel phenotype observed in CS individuals (Goodwin, Tidyman, et al., 2014). Although these mouse models have provided insight into the features of CS, more work is necessary to understand the underlying pathogenesis of CS.
CFC is a multiple congenital anomaly disorder characterized by craniofacial dysmorphia including macrocephaly, bitemporal narrowing, convex facial profile and hypoplastic supraorbital ridges, ectodermal abnormalities, congenital heart defects, growth delays, and neurocognitive deficits (Fig. 2E and F; Goodwin et al., 2013; Pierpont et al., 2014). CFC is caused by heterozygous, activating germline mutations in KRAS, BRAF, MAP2K1 (MEK1), or MAP2K2 (MEK2), all components of the RAS/MAPK pathway (Niihori et al., 2006; Rodriguez-Viciana et al., 2006). BRAF mutations have been found in human cancers, and the BRAFV600E mutation is the most commonly detected (Davies et al., 2002); the kinase activity of this mutation is 10- to 50-fold higher than that of other BRAF mutations responsible for CFC (Wan et al., 2004). The B-Raf+/LSLV600E mouse model expresses the constitutively active B-RafV600E allele but at 5–10% the level of the wild-type allele to model the BRAF activity in CFC individuals more closely. This model phenocopies features of CFC, including reduced size and body weight, more rounded and shorter heads due to changes in the shape of the frontal and parietal bones, cataracts, neurological defects including hyperactivity and seizures, and cardiomegaly due to an increase in the number of cardiomyocytes (Urosevic et al., 2011).
Additional mouse models of high- or intermediate-activity BRAF mutations have been developed. A conditional knock-in mouse for the intermediate-activity mutant BrafL597V phenocopies features of CFC including short stature, blunt nose, cardiac hypertrophy, and predisposition to develop benign tumors including papillomas and intestinal polyps (Andreadi et al., 2012). Another model expressing the BrafQ241R mutation, which is the most frequent mutation in the CFC population, manifests several features of CFC, including heart defects and craniofacial anomalies such as mandibular hypoplasia, before dying embryonically (Inoue et al., 2014). Also, CFC zebrafish models expressing a kinase-activating BRAFQ257R allele or kinase-inactivating BRAFG596V allele develop craniofacial anomalies (Anastasaki, Estep, Marais, Rauen, & Patton, 2009). These CFC models have elucidated the biological effects of specific BRAF mutations and revealed the importance of BRAF activity levels in CFC.
Drugs developed to target the RAS pathway in cancer have enormous potential for treatment of developmental syndromes caused by its dysregulation (Rauen et al., 2011, 2015). Utilizing animal models, researchers have begun testing the potential of RAS pathway inhibitors in treatment of the RASopathies. Treatment of NS mice with the MEK1/2 inhibitor PD0325901 restored their weight and length with partial improvement of the heart defects (Chen et al., 2010; Wu et al., 2011). Additionally, when NS mice were treated in utero with PD0325901, the craniofacial defects in the mice improved (Wu et al., 2011), and in utero treatment with U0126, an inhibitor of MEK1/2, resulted in complete rescue of the development and growth of the neural crest-derived bones of the face (Nakamura et al., 2007). In CS mice, the enamel defect was rescued in adult mice treated with PD0325901; however, treatment with the PI3K inhibitor GDC0941 had no effect on the disrupted ameloblasts or enamel defect, indicating that signaling through MEK is critical in enamel development (Goodwin, Tidyman, et al., 2014). Treatment of the CFC zebrafish model with low doses of PD0325901 at early stages of development ameliorated craniofacial defects (Anastasaki, Rauen, & Patton, 2012). Treatment of the BrafQ241R/+ embryos with PD0325091 rescued embryonic lethality and ameliorated edema and craniofacial defects, and furthermore, treatment in combination with a histone 3 demethylase inhibitor, GSK-J4, additionally rescued enlarged cardiac valves, suggesting the potential of combined therapies in treating the RASopathies (Inoue et al., 2014). Thus, drugs targeting the RAS pathway have potential in the treatment of RASopathies, but the complexity of RAS dysregulation in these syndromes means that further work will be required to determine optimal timing and dosage of drugs with specific targets.
4. CRANIOSYNOSTOSIS: PURSUING GENETIC AND PHARMACEUTICAL ALTERNATIVES TO SURGICAL TREATMENT
Craniosynostosis is the premature fusion of calvarial sutures, and multiple craniosynostosis syndromes, including Apert, Pfeiffer, and Crouzon, are caused by mutations in fibroblast growth factor receptors (FGFRs). Several mutations in FGFR1 and FGFR2 result in phenotypes that vary in severity yet share some characteristics. These mutations all result in gain of function of FGFR signaling, and a variety of specific molecular mechanisms have been shown to achieve this net effect. Mouse models have been invaluable in understanding how altered FGFR signaling contributes to the pathogenesis of these syndromes.
Apert syndrome is defined by craniosynostosis involving coronal sutures, midface hypoplasia, and bony and/or cutaneous syndactyly of the hands and feet (Bonaventure &El Ghouzzi, 2003; Wilkie, Oldridge, Tang, & Maxson, 2001). Individuals with Apert syndrome also have varying degrees of malformations of the nervous, respiratory, cardiovascular, and genitourinary systems. Furthermore, they have distinct craniofacial characteristics including increased incidence of cleft palate, narrow, high-arched palate and hypoplastic maxilla, hypodontia, delayed eruption, and dental crowding (Kreiborg & Cohen, 1992; Letra et al., 2007). Apert syndrome is caused by heterozygous, gain-of-function mutations affecting FGFR2, and more than 98% of individuals diagnosed with Apert syndrome carry a Ser252Trp (FGFR2 S252W; 66%) or Pro253Arg (FGFR2 P253R; 32%) mutation (Park et al., 1995; Wilkie et al., 1995). The altered amino acids lie in the linker region between the second and third extracellular immunoglobulin-like domains (IgII and IgIII) that mediate FGF ligand binding. The region encoding IgIII undergoes alternative splicing to form two well-described isoforms, FGFR2b and FGFR2c, which are expressed in the epithelium and mesenchyme, respectively, and have differing affinities for specific FGF ligands. Both mutations have been shown to increase FGFR2 ligand-binding affinity (Anderson, Burns, Enriquez-Harris, Wilkie, & Heath, 1998; Yu, Herr, Waksman, & Ornitz, 2000); the Ser252Trp mutation increases the affinity of FGFR2 for a subset of FGFs, whereas the Pro253Arg mutation increases affinity of FGFR2 for FGF indiscriminately (Ibrahimi et al., 2001).
Mouse models of the two most common Apert syndrome mutations in FGFR2 have shed light on the cellular mechanisms by which these mutations lead to syndromic phenotypes. The Fgfr2-Ser250Trp mutation resulted in mice that exhibit craniosynostosis, which is most pronounced in the coronal sutures, and skull malformations (Chen, Li, Li, Engel, & Deng, 2003; Wang et al., 2005). Increased Bax expression and apoptosis in the suture mesenchyme (Chen et al., 2003), as well as increased proliferation of chondrocytes and osteoblasts (Wang et al., 2005), have been suggested to contribute to craniosynostosis pathology in other mouse models of Apert syndrome.
When the P253R mutation was inserted in the Fgfr2 gene using a knock-in approach, the mice had premature closure of the coronal suture, as well as retarded growth of the synchondroses of the cranial base and growth plates of long bones, due to abnormalities in both osteogenesis and chondrogenesis (Wang et al., 2010; Yin et al., 2008). Of note, Fgfr2S250T mice did not develop syndactyly, whereas in rare instances Fgfr2P253R mice did. These data correlate with genotype–phenotype studies showing that the Pro253Arg mutation causes more severe syndactyly, whereas cleft palate is more common in Ser252Trp individuals (Slaney et al., 1996).
Furthermore, mutations causing ectopic expression of FGFR2 splice variants can result in increased ligand-dependent activation of the receptor. De novo Alu insertions within or upstream of alternatively spliced exon 9 (IIIC) of FGFR2 (Oldridge et al., 1999) resulted in ectopic expression of FGFR2b in mesenchymal tissues of the limb bud that normally express FGFR2c, suggesting a possible mechanism for the syndactyly phenotype in Apert syndrome. Similarly, excision of a single copy of FgfR2-IIIc in mice resulted in a gain-of-function mutation causing precocious ossification of the coronal sutures, zygomatic arch joints, and the sternebrae, as well as major defects in the kidney, lung, and lacrimal glands (Hajihosseini, Wilson, De Moerlooze, & Dickson, 2001). In this model, FgfR2-IIIb expression is substantially elevated in calvarial sutures and zygomatic joints, suggesting that cells in the sutures would respond to a broader array of FGF ligands and undergo differentiation prematurely, resulting in premature suture ossification. Thus, Apert syndrome appears to circumvent the biochemical and developmental regulatory mechanisms that are normally imposed by tissue-specific alternative splicing of Fgfr2, resulting in ectopic ligand-dependent receptor activation (Yu & Ornitz, 2001).
Similar to Apert syndrome, Pfeiffer syndrome is characterized by premature fusion of several cranial sutures, widely spaced eyes, a small nose, and midface hypoplasia (Pfeiffer, 1964; Vogels & Fryns, 2006). Additionally, individuals with Pfeiffer syndrome have hand and foot anomalies, including broad thumbs, cutaneous syndactly, shortened fingers, and medially deviated broad toes; however, unlike Apert syndrome, syndactyly is not a feature of Pfeiffer syndrome. Pfeiffer syndrome is caused by mutations in both FGFR1 and FGFR2, including a cytosine to guanine conversion in exon 5 of FGFR1 that results in a proline to arginine substitution in the extracellular domain (Muenke et al., 1994; Schell et al., 1995). When the Pro250Arg mutation was introduced into Fgfr1, mimicking the human mutation, the mice exhibited craniosynostosis, a dome-shaped skull, and a shortened snout (Zhou et al., 2000).
The phenotype of Crouzon syndrome is similar to that of Pfeiffer syndrome and includes craniosynostosis with abnormal skull shape and prominent eyes secondary to early fusion of the cranial sutures, without digital malformations like syndactyly (Crouzon, 1912). More than 30 different point mutations in the extracellular domain of FGFR2, most destroying or creating a cysteine residue, have been identified in individuals with Crouzon syndrome (Reardon et al., 1994; Wilkie et al., 1995). When FGFR2/Neu chimeras were generated with these mutations and expressed in NIH 3T3 cell cultures, FGFR2 was constitutively active due to aberrant intermolecular disulfide bonds (Galvin, Hart, Meyer, Webster, & Donoghue, 1996). These data suggest that the underlying pathology of Crouzon syndrome is the creation of unpaired cysteine residues, which facilitate the formation of intermolecular disulphide bonds, causing ligand-independent dimerization, phosphorylation, and signaling. The common Pfeiffer/Crouzon gain-of-function mutation Cys342Tyr was introduced into Fgfr2c, and the Fgfr2cC342Y/+ heterozygote mice were characterized by a shortened face, protruding eyes, and premature fusion of cranial sutures (Eswarakumar et al., 2004), due to ligand-independent activation of FGFR2 via disulfide bond dimerization (Eswarakumar et al., 2006). Notably, when a mutation inhibiting FRS2 binding was introduced together with the C342Y mutation, the phenotype was rescued, indicating that the FRS2 adaptor is a key mediator of FGFR2 signaling in craniosynostosis pathogenesis and suggesting a potential point of therapeutic intervention (Eswarakumar et al., 2006).
Currently, treatment of craniosynostosis is almost exclusively surgical and consists of removing fused sutures with osteotomies and reconstructing the skull (Panchal & Uttchin, 2003). More recently, surgeons have utilized distraction osteogenesis, in which osteotomies are performed and force is applied by an external or internal device to separate the skull fragments (Gasparini, Di Rocco, Tamburrini, & Pelo, 2012). Additionally, specific synostoses may be treated with minimally invasive endoscopic repairs that result in reduced blood loss and earlier hospital discharge ( Jimenez & Barone, 1998). Due to the morbidity associated with multiple surgeries to treat craniosynostosis, pharmacologic approaches are an attractive option to consider. Utilizing mouse models to understand the underlying pathogenesis of craniosynostosis associated with FGFR mutations has pinpointed downstream effectors that may serve as targets in treatment of FGFR-related craniosynostosis syndromes. Treatment of calvarial explants from Crouzon-like Fgfr2cC342Y/+ mice with the small-molecule FGFR inhibitors PLX052 or PD173074 prevented premature suture fusion (Eswarakumar et al., 2006; Perlyn, Morriss-Kay, Darvann, Tenenbaum, & Ornitz, 2006). Similarly, treatment of cultured calvaria from Apert-like P253R mice with the ERK1/2 inhibitor PD98059 partially alleviated the coronal suture fusion (Yin et al., 2008). In vivo, mice with the Fgfr2S252W mutation expressing a small hairpin RNA targeting Fgfr2S252W had patent cranial sutures and normal skull development (Shukla, Coumoul, Wang, Kim, & Deng, 2007). Furthermore, increased phosphorylation of ERK1/2 was observed in Fgfr2S252W mice, and treatment of these mice in utero at E18.5 and postnatally with U0126, an inhibitor of MEK1/2, resulted in rescue of the craniosynostosis phenotype (Shukla et al., 2007). Thus, utilization of mouse models has elucidated potential downstream targets of FGFR signaling for pharmacological treatment of craniosynostosis. However, resynostosis has been noted after withdrawal of inhibitors, and further investigation is necessary to develop drugs with specific targets to maximize efficacy and minimize side effects.
5. XLHED: DEVELOPING TREATMENT BASED ON KNOWLEDGE GAINED FROM MOUSE AND CANINE MODELS
In 1848, an early case study reported a male subject with an “almost complete absence of hair; the teeth being not more than four in number; the delicate structure of the skin; and the absence of sensible perspiration and tears” (Thurnam, 1848). In 1875, Charles Darwin made similar observations on a family of males in India (Darwin, 1875). These men suffered from a hereditary condition that disrupts the development of ectodermal structures and their appendages, termed ectodermal dysplasia (ED; Smith, 1929).
Over 150 syndromes have been categorized as EDs, and the most prevalent subgroup is comprised of the hypohidrotic ectodermal dysplasias (HEDs), which affect more than 1 in 17,000 live births (Itin & Fistarol, 2004; Pinheiro & Freire-Maia, 1994). Clinical features of HED include sparseness of scalp and body hair (hypotrichosis); reduced sweating (hypohidrosis) and tear production; and malformation and reduced number of teeth (hypodontia), with an average of nine permanent teeth (Fig. 3; Clarke, 1987; Kobielak et al., 2001; Lexner, Bardow, Hertz, Nielsen, & Kreiborg, 2007; Solomon & Keuer, 1980). In addition, many HED patients exhibit abnormalities in facial appearance, such as prominent forehead, depressed nasal bridge, hypoplastic nose, prominent supraorbital ridges, and thick lips (Goodwin, Larson, et al., 2014; Gunduz Arslan, Devecioglu Kama, Ozer, & Yavuz, 2007;Johnson et al., 2002; Lexner et al., 2007). These clinical phenotypes serve as the initial diagnostic features for most affected individuals with HED. Early diagnosis is crucial because affected children can develop recurrent hyperpyrexia due to impaired sweating and temperature control ability, which can result in febrile seizures; some authors have suggested that neurologic damage can result and have proposed a mortality rate of about 30% during the first 2 years of life (Clarke, 1987; Salisbury & Stothers, 1981).
Figure 3.
Craniofacial and dental characteristics of XLHED. (A) 13-year-old male with facial characteristics of XLHED including short face with proportionally longer chin and midface, narrow and pointed nose, narrow mouth with full, rounded lower lip and (B) dental features such as missing teeth and conical-shaped incisors and molars with abnormal cuspal morphology. (C) A 21-year-old male with midface hypoplasia and prognathic mandible and chin and short philtrum, and (D) missing teeth, typical of XLHED.
HED individuals carry mutations in one or more of the core genes in the ectodysplasin pathway: ectodysplasin ligand (EDA), ectodysplasin receptor (EDAR), or EDAR-associated death domain adaptor protein (EDARADD; reviewed in Sadier, Viriot, Pantalacci, & Laudet 2014). EDA is a type II transmembrane tumor necrosis factor superfamily member that is proteolytically processed into a soluble trimeric form to bind the receptor, EDAR. Upon activation, EDAR recruits EDARADD via its intracellular death domain region. In turn, EDARADD transduces the signal to activate the NF-κB pathway, which mediates the transcriptional activation of the target genes involved in hair, sweat gland, and tooth development (Lefebvre, Fliniaux, Schneider, & Mikkola, 2012). In rare cases, HED can be associated with immune deficiency caused by dysregulation of NF-κB (Doffinger et al., 2001; Jain et al., 2001; Zonana et al., 2000). EDA has several splicing isoforms; EDA-A1 and -A2 interact with EDAR and EDA2R (formerly known as XEDAR), respectively.
Mutations in EDAR and EDARADD are inherited in an autosomal dominant or recessive fashion, whereas EDA mutations are inherited in an X-linked manner. Phenotypes caused by mutations at different points along the EDA pathway are clinically similar, with autosomal dominant forms of EDAR and EDARADD exhibiting milder features. Here, we will focus on X-linked HED (XLHED), which is caused by mutations in EDA and is the most common form of HED.
Tabby mice that exhibit clinical features of XLHED were described in the mid-1900s, but it was not until three decades later that the Tabby gene was confirmed to be orthologous to human EDA through synteny mapping and comparison of predicted amino sequence (Bayes et al., 1998; Falconer, 1952; Ferguson et al., 1997; Srivastava et al., 1997). Tabby mice carry spontaneous loss-of-function mutations in Eda, resulting in phenotypes comparable to those observed in XLHED individuals: hypodontia with incomplete penetrance; reduced size and cusps of the molars; reduction in four pelage hair types (awl, auchene, guard, zigzag) to a single awl-like hair type; missing hair on tail and retroauricular regions; missing sweat glands on footpads; and reduction in size of various glands, including lacrimal and salivary glands (Cui et al., 2003; Gruneberg, 1965, 1971; Pispa et al., 1999; Srivastava et al., 1997). In contrast, overexpression of the Eda-A1 transgene in developing murine ectoderm resulted in supernumerary mammary glands and teeth, hypertrophy of sebaceous glands, and abnormal composition and structure of pelage hairs (Mustonen et al., 2003; Srivastava et al., 2001). These mouse models with dysregulation of the EDA pathway suggest the importance of the EDA pathway in fine-tuning the size, number, and morphology of ectodermal organs.
Controlled expression of a tetracycline-regulated Eda-A1 transgene during Tabby embryogenesis revealed that ectodysplasin signaling is required for hair development within defined spatiotemporal windows (Cui, Kunisada, Esibizione, Douglass, & Schlessinger, 2009). Tabby phenotypes can also be ameliorated by administration of recombinant Fc:EDA1, which contains the EDAR-binding domain linked to the IgG1 Fc domain to allow trafficking across the placental barrier and trimerization of the ligand (Fig. 4; Gaide & Schneider, 2003). As with transgenic rescue, Fc:EDA1 efficacy was timing dependent; fetal, but not neonatal, administration rescued tooth morphology, as well as some hair and gland phenotypes. Interestingly, whereas transgenic expression of Eda-A1 was able to rescue the number but not the morphology of molars, Fc:EDA1 treatment restored morphology but not molar tooth number (Gaide & Schneider, 2003).
Figure 4.
Schematic of recombinant EDA therapy. In unaffected individuals, EDA binds EDAR, activating downstream signaling that results in tooth, hair, and sweat gland development. XLHED individuals are missing EDA, which results in missing and conical-shaped teeth, sparse, thin hair, and hypoplastic sweat glands. EDI200 therapy replaces missing EDA in XLHED and rescues tooth, hair, and sweat gland development.
Subsequently, a colony of dogs with XLHED-like features was characterized and found to carry a spontaneous mutation in Eda that resulted in truncated EDA-A1 and -A2 (Casal, Jezyk, Greek, Goldschmidt, & Patterson, 1997). EDA replacement experiments were then performed in the canine model, but because immunoglobulins are not transferred transplacentally in dogs, Fc:EDA1 was administered postnatally via intravenous injection. The treatment effectively rescued the sweat glands, as observed in Tabby mice. Although hair growth was not improved, the number and shape of permanent teeth were restored in most of the treated dogs (Casal et al., 2007). Furthermore, susceptibility to eye and respiratory infections was also reduced through restoration of lacrimal as well as tracheal and bronchial glands. These results collectively demonstrate that short-term treatment with recombinant EDA during a therapeutic window can dramatically improve the XLHED phenotype in murine and canine models. Importantly, the treatment did not cause any noticeable side effects during or after the injections, showing great promise for the potential utility of such a therapeutic intervention in patients with XLHED. Currently, clinical trials are underway to administer recombinant EDA to XLHED-affected males in the immediate postnatal period (Fig. 4), constituting the first attempt to treat a structural birth defect in humans with a targeted drug therapy.
6. CONCLUDING THOUGHTS
The conditions that we have reviewed in this chapter, including orofacial clefting, the RASopathies, craniosynostosis syndromes, and XLHED, illustrate the exciting recent advances in our understanding of the molecular underpinnings of craniofacial conditions. These disorders lie at various points along a spectrum that has at one end basic discovery science and at the other end clinical trials of targeted therapies. The studies described above have led to an increased understanding of the etiology of these conditions and have revealed the pathways and processes underlying craniofacial development as well as general principles that are applicable to our broader understanding of organogenesis. These examples provide hope that, as the field of craniofacial biology continues to move forward, a shift will ensue from surgical correction toward medical treatment and even prevention. The barriers to such a goal include the timing of diagnosis, developmental pleiotropy of signaling pathways, and the unpredictable consequences of specific gene mutations in a given individual. Overcoming these barriers will require continued intensive study of the basic mechanisms that underlie these diseases.
ACKNOWLEDGMENTS
The authors were funded by the following NIDCR grants: U01-DE024440 and R01-DE021420 to O.D.K., R01-DE023337 to J.O.B., and F30-DE025160 to R.K.
REFERENCES
- Alappat S, Zhang ZY, & Chen YP (2003). Msx homeobox gene family and craniofacial development. Cell Research, 13, 429–442. [DOI] [PubMed] [Google Scholar]
- Anastasaki C, Estep AL, Marais R, Rauen KA, & Patton EE (2009). Kinase-activating and kinase-impaired cardio-facio-cutaneous syndrome alleles have activity during zebrafish development and are sensitive to small molecule inhibitors. Human Molecular Genetics, 18, 2543–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasaki C, Rauen KA, & Patton EE (2012). Continual low-level MEK inhibition ameliorates cardio-facio-cutaneous phenotypes in zebrafish. Disease Models & Mechanisms, 5, 546–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J, Burns HD, Enriquez-Harris P, Wilkie AO, & Heath JK (1998). Apert syndrome mutations in fibroblast growth factor receptor 2 exhibit increased affinity for FGF ligand. Human Molecular Genetics, 7, 1475–1483. [DOI] [PubMed] [Google Scholar]
- Andreadi C, Cheung LK, Giblett S, Patel B, Jin H, Mercer K, et al. (2012). The intermediate-activity (L597V)BRAF mutant acts as an epistatic modifier of oncogenic RAS by enhancing signaling through the RAF/MEK/ERK pathway. Genes & Development, 26, 1945–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y, Niihori T, Kawame H, Kurosawa K, Ohashi H, Tanaka Y, et al. (2005). Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nature Genetics, 37, 1038–1040. [DOI] [PubMed] [Google Scholar]
- Araki T, Mohi MG, Ismat FA, Bronson RT, Williams IR, Kutok JL, et al. (2004). Mouse model of Noonan syndrome reveals cell type- and gene dosage-dependent effects of Ptpn11 mutation. Nature Medicine, 10, 849–857. [DOI] [PubMed] [Google Scholar]
- Bayes M, Hartung AJ, Ezer S, Pispa J, Thesleff I, Srivastava AK, et al. (1998). The anhidrotic ectodermal dysplasia gene (EDA) undergoes alternative splicing and encodes ectodysplasin-A with deletion mutations in collagenous repeats. Human Molecular Genetics, 7, 1661–1669. [DOI] [PubMed] [Google Scholar]
- Beaty TH, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB, et al. (2010). A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nature Genetics, 42, 525–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty TH, Ruczinski I, Murray JC, Marazita ML, Munger RG, Hetmanski JB, et al. (2011). Evidence for gene-environment interaction in a genome wide study of nonsyndromic cleft palate. Genetic Epidemiology, 35, 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertuzzi S, Hindges R, Mui SH, O’Leary DD, & Lemke G (1999). The homeodomain protein vax1 is required for axon guidance and major tract formation in the developing forebrain. Genes & Development, 13, 3092–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W, Huang W, Whitworth DJ, Deng JM, Zhang Z, Behringer RR, et al. (2001). Haploinsufficiency of Sox9 results in defective cartilage primordia and premature skeletal mineralization. Proceedings of the National Academy of Sciences of the United States of America, 98, 6698–6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum S, Ludwig KU, Reutter H, Herms S, Steffens M, Rubini M, et al. (2009). Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nature Genetics, 41, 473–477. [DOI] [PubMed] [Google Scholar]
- Bonaventure J, & El Ghouzzi V (2003). Molecular and cellular bases of syndromic craniosynostoses. Expert Reviews in Molecular Medicine, 5, 1–17. [DOI] [PubMed] [Google Scholar]
- Bowen JM, & Connolly HM (2014). Of Marfan’s syndrome, mice, and medications. The New England Journal of Medicine, 371, 2127–2128. [DOI] [PubMed] [Google Scholar]
- Brewer S, Feng W, Huang J, Sullivan S, & Williams T (2004). Wnt1-Cre-mediated deletion of AP-2alpha causes multiple neural crest-related defects. Developmental Biology, 267, 135–152. [DOI] [PubMed] [Google Scholar]
- Britanova O, Depew MJ, Schwark M, Thomas BL, Miletich I, Sharpe P, et al. (2006). Satb2 haploinsufficiency phenocopies 2q32-q33 deletions, whereas loss suggests a fundamental role in the coordination of jaw development. American Journal of Human Genetics, 79, 668–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush JO, & Jiang R (2012). Palatogenesis: Morphogenetic and molecular mechanisms of secondary palate development. Development, 139, 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush JO, & Soriano P (2010). Ephrin-B1 forward signaling regulates craniofacial morphogenesis by controlling cell proliferation across Eph-ephrin boundaries. Genes & Development, 24, 2068–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal ML, Jezyk PF, Greek JM, Goldschmidt MH, & Patterson DF (1997). X-linked ectodermal dysplasia in the dog. The Journal of Heredity, 88, 513–517. [DOI] [PubMed] [Google Scholar]
- Casal ML, Lewis JR, Mauldin EA, Tardivel A, Ingold K, Favre M, et al. (2007). Significant correction of disease after postnatal administration of recombinant ectodysplasin A in canine X-linked ectodermal dysplasia. American Journal of Human Genetics, 81, 1050–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Li D, Li C, Engel A, & Deng CX (2003). A Ser252Trp [corrected] substitution in mouse fibroblast growth factor receptor 2 (Fgfr2) results in craniosynostosis. Bone, 33, 169–178. [DOI] [PubMed] [Google Scholar]
- Chen X, Mitsutake N, LaPerle K, Akeno N, Zanzonico P, Longo VA, et al. (2009). Endogenous expression of Hras(G12V) induces developmental defects and neoplasms with copy number imbalances of the oncogene. Proceedings of the National Academy of Sciences of the United States of America, 106, 7979–7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PC, Wakimoto H, Conner D, Araki T, Yuan T, Roberts A, et al. (2010). Activation of multiple signaling pathways causes developmental defects in mice with a Noonan syndrome-associated Sos1 mutation. The Journal of Clinical Investigation, 120, 4353–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu HH, Wu MH, Wang JK, Lu CW, Chiu SN, Chen CA, et al. (2013). Losartan added to beta-blockade therapy for aortic root dilation in Marfan syndrome: A randomized, open-label pilot study. Mayo Clinic Proceedings, 88, 271–276. [DOI] [PubMed] [Google Scholar]
- Cirstea IC, Kutsche K, Dvorsky R, Gremer L, Carta C, Horn D, et al. (2010). A restricted spectrum of NRAS mutations causes Noonan syndrome. Nature Genetics, 42, 27–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke A (1987). Hypohidrotic ectodermal dysplasia. Journal of Medical Genetics, 24, 659–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins E, & Turner G (1973). The Noonan syndrome—A review of the clinical and genetic features of 27 cases. The Journal of Pediatrics, 83, 941–950. [DOI] [PubMed] [Google Scholar]
- Compagni A, Logan M, Klein R, & Adams RH (2003). Control of skeletal patterning by ephrinB1-EphB interactions. Developmental Cell, 5, 217–230. [DOI] [PubMed] [Google Scholar]
- Cordeddu V, Di Schiavi E, Pennacchio LA, Ma’ayan A, Sarkozy A, Fodale V, et al. (2009). Mutation of SHOC2 promotes aberrant protein N-myristoylation and causes Noonan-like syndrome with loose anagen hair. Nature Genetics, 41, 1022–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouzon O (1912). Dysostose cranio-faciale hereditaire. Bulletins et mémoires de la Société médicale des hôpitaux de Paris, 33, 545–555. [Google Scholar]
- Cui CY, Durmowicz M, Ottolenghi C, Hashimoto T, Griggs B, Srivastava AK, et al. (2003). Inducible mEDA-A1 transgene mediates sebaceous gland hyperplasia and differential formation of two types of mouse hair follicles. Human Molecular Genetics, 12, 2931–2940. [DOI] [PubMed] [Google Scholar]
- Cui CY, Kunisada M, Esibizione D, Douglass EG, & Schlessinger D (2009). Analysis of the temporal requirement for eda in hair and sweat gland development. Journal of Investigative Dermatology, 129(4), 984–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C (1875). Insectivorous Plants. The variations of animals and plants under domestication: Vol. 2 (p. 319). London: John Murray. [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. (2002). Mutations of the BRAF gene in human cancer. Nature, 417, 949–954. [DOI] [PubMed] [Google Scholar]
- De Felice M, Ovitt C, Biffali E, Rodriguez-Mallon A, Arra C, Anastassiadis K, et al. (1998). A mouse model for hereditary thyroid dysgenesis and cleft palate. Nature Genetics, 19, 395–398. [DOI] [PubMed] [Google Scholar]
- Dietz HC, Cutting GR, Pyeritz RE, Maslen CL, Sakai LY, Corson GM, et al. (1991). Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature, 352, 337–339. [DOI] [PubMed] [Google Scholar]
- Ding H, Wu X, Bostrom H, Kim I, Wong N, Tsoi B, et al. (2004). A specific requirement for PDGF-C in palate formation and PDGFR-alpha signaling. Nature Genetics, 36, 1111–1116. [DOI] [PubMed] [Google Scholar]
- Dixon J, Jones NC, Sandell LL, Jayasinghe SM, Crane J, Rey JP, et al. (2006). Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proceedings of the National Academy of Sciences of the United States of America, 103, 13403–13408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MJ, Marazita ML, Beaty TH, & Murray JC (2011). Cleft lip and palate: Understanding genetic and environmental influences. Nature Reviews. Genetics, 12, 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J, Trainor P, & Dixon MJ (2007). Treacher Collins syndrome. Orthodontics & Craniofacial Research, 10, 88–95. [DOI] [PubMed] [Google Scholar]
- Doffinger R, Smahi A, Bessia C, Geissmann F, Feinberg J, Durandy A, et al. (2001). X-linked anhidrotic ectodermal dysplasia with immunodeficiency is caused by impaired NF-kappaB signaling. Nature Genetics, 27, 277–285. [DOI] [PubMed] [Google Scholar]
- Dudas M, Kim J, Li WY, Nagy A, Larsson J, Karlsson S, et al. (2006). Epithelial and ectomesenchymal role of the type I TGF-beta receptor ALK5 during facial morphogenesis and palatal fusion. Developmental Biology, 296, 298–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SJ, Fowlie A, Cust MP, Liu DT, Young ID, & Dixon MJ (1996). Prenatal diagnosis in Treacher Collins syndrome using combined linkage analysis and ultrasound imaging. Journal of Medical Genetics, 33, 603–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estep AL, Tidyman WE, Teitell MA, Cotter PD, & Rauen KA (2006). HRAS mutations in Costello syndrome: Detection of constitutional activating mutations in codon 12 and 13 and loss of wild-type allele in malignancy. American Journal of Medical Genetics. Part A, 140, 8–16. [DOI] [PubMed] [Google Scholar]
- Eswarakumar VP, Horowitz MC, Locklin R, Morriss-Kay GM, &Lonai P (2004). A gain-of-function mutation of Fgfr2c demonstrates the roles of this receptor variant in osteogenesis. Proceedings of the National Academy of Sciences of the United States of America, 101, 12555–12560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswarakumar VP, Ozcan F, Lew ED, Bae JH, Tome F, Booth CJ, et al. (2006). Attenuation of signaling pathways stimulated by pathologically activated FGF-receptor 2 mutants prevents craniosynostosis. Proceedings of the National Academy of Sciences of the United States of America, 103, 18603–18608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer DS (1952). A totally sex-linked gene in the house mouse. Nature, 169, 664–665. [DOI] [PubMed] [Google Scholar]
- Fazen LE, Elmore J, & Nadler HL (1967). Mandibulofacial dysostosis (Treacher-Collins syndrome). American Journal of Diseases of Children, 113, 405–410. [DOI] [PubMed] [Google Scholar]
- Ferguson BM, Brockdorff N, Formstone E, Ngyuen T, Kronmiller JE,& Zonana J (1997). Cloning of Tabby, the murine homolog of the human EDA gene: Evidence for a membrane-associated protein with a short collagenous domain. Human Molecular Genetics, 6, 1589–1594. [DOI] [PubMed] [Google Scholar]
- Ferone G, Thomason HA, Antonini D, De Rosa L, Hu B, Gemei M, et al. (2012). Mutant p63 causes defective expansion of ectodermal progenitor cells and impaired FGF signalling in AEC syndrome. EMBO Molecular Medicine, 4, 192–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaide O, & Schneider P (2003). Permanent correction of an inherited ectodermal dysplasia with recombinant EDA. Nature Medicine, 9, 614–618. [DOI] [PubMed] [Google Scholar]
- Galvin BD, Hart KC, Meyer AN, Webster MK, &Donoghue DJ (1996). Constitutive receptor activation by Crouzon syndrome mutations in fibroblast growth factor receptor (FGFR)2 and FGFR2/Neu chimeras. Proceedings of the National Academy of Sciences of the United States of America, 93, 7894–7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparini G, Di Rocco C, Tamburrini G, & Pelo S (2012). External craniofacial osteodistraction in complex craniosynostoses. Child’s Nervous System: ChNS: Official Journal of the International Society for Pediatric Neurosurgery, 28, 1565–1570. [DOI] [PubMed] [Google Scholar]
- Goodwin AF, Larson JR, Jones KB, Liberton DK, Landan M, Wang Z, et al. (2014). Craniofacial morphometric analysis of individuals with X-linked hypohidrotic ectodermal dysplasia. Molecular Genetics & Genomic Medicine, 2, 422–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin AF, Oberoi S, Landan M, Charles C, Groth J, Martinez A, et al. (2013). Craniofacial and dental development in cardio-facio-cutaneous syndrome: The importance of Ras signaling homeostasis. Clinical Genetics, 83, 539–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin AF, Oberoi S, Landan M, Charles C, Massie JC, Fairley C, et al. (2014). Craniofacial and dental development in Costello syndrome. American Journal of Medical Genetics. Part A, 164A, 1425–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin AF, Tidyman WE, Jheon AH, Sharir A, Zheng X, Charles C, et al. (2014). Abnormal Ras signaling in Costello syndrome (CS) negatively regulates enamel formation. Human Molecular Genetics, 23, 682–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant SF, Wang K, Zhang H, Glaberson W, Annaiah K, Kim CE, et al. (2009). A genome-wide association study identifies a locus for nonsyndromic cleft lip with or without cleft palate on 8q24. The Journal of Pediatrics, 155, 909–913. [DOI] [PubMed] [Google Scholar]
- Green RM, Feng W, Phang T, Fish JL, Li H, Spritz RA, et al. (2015). Tfap2a-dependent changes in mouse facial morphology result in clefting that can be ameliorated by a reduction in Fgf8 gene dosage. Disease Models & Mechanisms, 8, 31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritli-Linde A (2008). The etiopathogenesis of cleft lip and cleft palate: Usefulness and caveats of mouse models. Current Topics in Developmental Biology, 84, 37–138. [DOI] [PubMed] [Google Scholar]
- Groenink M, den Hartog AW, Franken R, Radonic T, de Waard V, Timmermans J, et al. (2013). Losartan reduces aortic dilatation rate in adults with Marfan syndrome: A randomized controlled trial. European Heart Journal, 34, 3491–3500. [DOI] [PubMed] [Google Scholar]
- Grosen D, Bille C, Pedersen JK, Skytthe A, Murray JC, & Christensen K (2010). Recurrence risk for offspring of twins discordant for oral cleft: A population-based cohort study of the Danish 1936-2004 cleft twin cohort. American Journal of Medical Genetics. Part A, 152A, 2468–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruneberg H (1965). Genes and genotypes affecting the teeth of the mouse. Journal of Embryology and Experimental Morphology, 14, 137–159. [PubMed] [Google Scholar]
- Gruneberg H (1971). The tabby syndrome in the mouse. Proceedings of the Royal Society of London, Series B: Biological Sciences, 179, 139–156. [DOI] [PubMed] [Google Scholar]
- Gunduz Arslan S, Devecioglu Kama J, Ozer T, & Yavuz I (2007). Craniofacial and upper airway cephalometrics in hypohidrotic ectodermal dysplasia. Dento Maxillo Facial Radiology, 36, 478–483. [DOI] [PubMed] [Google Scholar]
- Habashi JP, Judge DP, Holm TM, Cohn RD, Loeys BL, Cooper TK, et al. (2006). Losartan, an AT1 antagonist, prevents aortic aneurysm in a mouse model of Marfan syndrome. Science, 312, 117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajihosseini MK, Wilson S, De Moerlooze L, & Dickson C (2001). A splicing switch and gain-of-function mutation in FgfR2-IIIc hemizygotes causes Apert/Pfeiffer-syndrome-like phenotypes. Proceedings of the National Academy of Sciences of the United States of America, 98, 3855–3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Mayo J, Xu X, Li J, Bringas P Jr., Maas RL, et al. (2009). Indirect modulation of Shh signaling by Dlx5 affects the oral-nasal patterning of palate and rescues cleft palate in Msx1-null mice. Development, 136, 4225–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart AW, Morgan JE, Schneider J, West K, McKie L,Bhattacharya S, et al. (2006). Cardiac malformations and midline skeletal defects in mice lacking filamin A. Human Molecular Genetics, 15, 2457–2467. [DOI] [PubMed] [Google Scholar]
- Hernandez-Porras I, Fabbiano S, Schuhmacher AJ, Aicher A, Canamero M, Camara JA, et al. (2014). K-RasV14I recapitulates Noonan syndrome in mice. Proceedings of the National Academy of Sciences of the United States of America, 111, 16395–16400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa R, Deng X, Takamori K, Xu X, Urata M, Bringas P Jr., et al. (2009). Epithelial-specific requirement of FGFR2 signaling during tooth and palate development. Journal of Experimental Zoology. Part B, Molecular and Developmental Evolution, 312B, 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Goudy SL, Ketova T, Litingtung Y, & Chiang C (2008). Gli3-deficient mice exhibit cleft palate associated with abnormal tongue development. Developmental Dynamics: An Official Publication of the American Association of Anatomists, 237, 3079–3087. [DOI] [PubMed] [Google Scholar]
- Ibrahimi OA, Eliseenkova AV, Plotnikov AN, Yu K, Ornitz DM, & Mohammadi M (2001). Structural basis for fibroblast growth factor receptor 2 activation in Apert syndrome. Proceedings of the National Academy of Sciences of the United States of America, 98, 7182–7187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa E, Watanabe A, Nakano Y, Akita S, Hirano A, Kinoshita A, et al. (2006). PAX9 and TGFB3 are linked to susceptibility to nonsyndromic cleft lip with or without cleft palate in the Japanese: Population-based and family-based candidate gene analyses. Journal of Human Genetics, 51, 38–46. [DOI] [PubMed] [Google Scholar]
- Iida K, Koseki H, Kakinuma H, Kato N, Mizutani-Koseki Y, Ohuchi H, et al. (1997). Essential roles of the winged helix transcription factor MFH-1 in aortic arch patterning and skeletogenesis. Development, 124, 4627–4638. [DOI] [PubMed] [Google Scholar]
- Ingraham CR, Kinoshita A, Kondo S, Yang B, Sajan S, Trout KJ, et al. (2006). Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6). Nature Genetics, 38, 1335–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Moriya M, Watanabe Y, Miyagawa-Tomita S, Niihori T, Oba D, et al. (2014). New BRAF knockin mice provide a pathogenetic mechanism of developmental defects and a therapeutic approach in cardio-facio-cutaneous syndrome. Human Molecular Genetics, 23, 6553–6566. [DOI] [PubMed] [Google Scholar]
- Ishii M, Han J, Yen HY, Sucov HM, Chai Y, & Maxson RE Jr. (2005). Combined deficiencies of Msx1 and Msx2 cause impaired patterning and survival of the cranial neural crest. Development, 132, 4937–4950. [DOI] [PubMed] [Google Scholar]
- Itin PH, & Fistarol SK (2004). Ectodermal dysplasias. American Journal of Medical Genetics. Part C, Seminars in Medical Genetics, 131C, 45–51. [DOI] [PubMed] [Google Scholar]
- Ito Y, Yeo JY, Chytil A, Han J, Bringas P Jr., Nakajima A, et al. (2003). Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development, 130, 5269–5280. [DOI] [PubMed] [Google Scholar]
- Iwata J, Hacia JG, Suzuki A, Sanchez-Lara PA, Urata M, & Chai Y (2012). Modulation of noncanonical TGF-beta signaling prevents cleft palate in Tgfbr2 mutant mice. The Journal of Clinical Investigation, 122, 873–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata J, Parada C, & Chai Y (2011). The mechanism of TGF-beta signaling during palate development. Oral Diseases, 17, 733–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata J, Suzuki A, Pelikan RC, Ho TV, Sanchez-Lara PA, Urata M, et al. (2013). Smad4-Irf6 genetic interaction and TGFbeta-mediated IRF6 signaling cascade are crucial for palatal fusion in mice. Development, 140, 1220–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Ma CA, Liu S, Brown M, Cohen J, & Strober W (2001). Specific missense mutations in NEMO result in hyper-IgM syndrome with hypohydrotic ectodermal dysplasia. Nature Immunology, 2, 223–228. [DOI] [PubMed] [Google Scholar]
- Jerome LA, & Papaioannou VE (2001). DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nature Genetics, 27, 286–291. [DOI] [PubMed] [Google Scholar]
- Jezewski PA, Vieira AR, Nishimura C, Ludwig B, Johnson M, O’Brien SE, et al. (2003). Complete sequencing shows a role for MSX1 in non-syndromic cleft lip and palate. Journal of Medical Genetics, 40, 399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Bush JO, & Lidral AC (2006). Development of the upper lip: Morphogenetic and molecular mechanisms. Developmental Dynamics: An Official Publication of the American Association of Anatomists, 235, 1152–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez DF, & Barone CM (1998). Endoscopic craniectomy for early surgical correction of sagittal craniosynostosis. Journal of Neurosurgery, 88, 77–81. [DOI] [PubMed] [Google Scholar]
- Johnson EL, Roberts MW, Guckes AD, Bailey LJ, Phillips CL, & Wright JT (2002). Analysis of craniofacial development in children with hypohidrotic ectodermal dysplasia. American Journal of Medical Genetics, 112, 327–334. [DOI] [PubMed] [Google Scholar]
- Jones NC, Lynn ML, Gaudenz K, Sakai D, Aoto K, Rey JP, et al. (2008). Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nature Medicine, 14, 125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jugessur A, Farlie PG, & Kilpatrick N (2009). The genetics of isolated orofacial clefts: From genotypes to subphenotypes. Oral Diseases, 15, 437–453. [DOI] [PubMed] [Google Scholar]
- Juriloff DM, & Fraser FC (1980). Genetic maternal effects on cleft lip frequency in A/J and CL/Fr mice. Teratology, 21, 167–175. [DOI] [PubMed] [Google Scholar]
- Kaartinen V, Voncken JW, Shuler C, Warburton D, Bu D, Heisterkamp N, et al. (1995). Abnormal lung development and cleft palate in mice lacking TGF-beta 3 indicates defects of epithelial-mesenchymal interaction. Nature Genetics, 11, 415–421. [DOI] [PubMed] [Google Scholar]
- Kim S, Lewis AE, Singh V, Ma X, Adelstein R, & Bush JO (2015). Convergence and extrusion are required for normal fusion of the mammalian secondary palate. PLoS Biology, 13(4), e1002122 10.1371/journal.pbio.1002122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobielak K, Kobielak A, Roszkiewicz J, Wierzba J, Limon J, & Trzeciak WH (2001). Mutations in the EDA gene in three unrelated families reveal no apparent correlation between phenotype and genotype in the patients with an X-linked anhidrotic ectodermal dysplasia. American Journal of Medical Genetics, 100, 191–197. [PubMed] [Google Scholar]
- Kohama K, Nonaka K, Hosokawa R, Shum L, & Ohishi M (2002). TGF-beta-3 promotes scarless repair of cleft lip in mouse fetuses. Journal of Dental Research, 81, 688–694. [DOI] [PubMed] [Google Scholar]
- Kondo S, Schutte BC, Richardson RJ, Bjork BC, Knight AS, Watanabe Y, et al. (2002). Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nature Genetics, 32, 285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiborg S, & Cohen MM Jr. (1992). The oral manifestations of Apert syndrome. Journal of Craniofacial Genetics and Developmental Biology, 12, 41–48. [PubMed] [Google Scholar]
- Lacro RV, Dietz HC, Sleeper LA, Yetman AT, Bradley TJ, Colan SD, et al. (2014). Atenolol versus losartan in children and young adults with Marfan’s syndrome. The New England Journal of Medicine, 371, 2061–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre S, Fliniaux I, Schneider P, & Mikkola ML (2012). Identification of ectodysplasin target genes reveals the involvement of chemokines in hair development. The Journal of Investigative Dermatology, 132, 1094–1102. [DOI] [PubMed] [Google Scholar]
- Leslie EJ, Mansilla MA, Biggs LC, Schuette K, Bullard S, Cooper M, et al. (2012). Expression and mutation analyses implicate ARHGAP29 as the etiologic gene for the cleft lip with or without cleft palate locus identified by genome-wide association on chromosome 1p22. Birth Defects Research. Part A, Clinical and Molecular Teratology, 94, 934–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letra A, de Almeida AL, Kaizer R, Esper LA, Sgarbosa S, & Granjeiro JM (2007). Intraoral features of Apert’s syndrome. Oral Surgery, Oral Medicine, Oral Pathology, Oral Radiology, and Endodontics, 103, e38–e41. [DOI] [PubMed] [Google Scholar]
- Levine AJ (1997). p53, the cellular gatekeeper for growth and division. Cell, 88, 323–331. [DOI] [PubMed] [Google Scholar]
- Lexner MO, Bardow A, Hertz JM, Nielsen LA, &Kreiborg S (2007). Anomalies of tooth formation in hypohidrotic ectodermal dysplasia. International Journal of Paediatric Dentistry/The British Paedodontic Society [and] the International Association of Dentistry for Children, 17, 10–18. [DOI] [PubMed] [Google Scholar]
- Li Y, Lacerda DA, Warman ML, Beier DR, Yoshioka H, Ninomiya Y, et al. (1995). A fibrillar collagen gene, Col11a1, is essential for skeletal morphogenesis. Cell, 80, 423–430. [DOI] [PubMed] [Google Scholar]
- Li SW, Prockop DJ, Helminen H, Fassler R, Lapvetelainen T, Kiraly K, et al. (1995). Transgenic mice with targeted inactivation of the Col2 alpha 1 gene for collagen II develop a skeleton with membranous and periosteal bone but no endochondral bone. Genes & Development, 9, 2821–2830. [DOI] [PubMed] [Google Scholar]
- Lidral AC, Romitti PA,Basart AM, Doetschman T, Leysens NJ, Daack-Hirsch S, et al. (1998). Association of MSX1 and TGFB3 with nonsyndromic clefting in humans. American Journal of Human Genetics, 63, 557–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Sun X, Braut A, Mishina Y, Behringer RR, Mina M, et al. (2005). Distinct functions for Bmp signaling in lip and palate fusion in mice. Development, 132, 1453–1461. [DOI] [PubMed] [Google Scholar]
- Loeys BL, Chen J, Neptune ER, Judge DP, Podowski M, Holm T, et al. (2005). A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFBR1 or TGFBR2. Nature Genetics, 37, 275–281. [DOI] [PubMed] [Google Scholar]
- Mangold E, Ludwig KU, Birnbaum S, Baluardo C, Ferrian M, Herms S, et al. (2010). Genome-wide association study identifies two susceptibility loci for non-syndromic cleft lip with or without cleft palate. Nature Genetics, 42, 24–26. [DOI] [PubMed] [Google Scholar]
- Mangold E, Ludwig KU, & Nothen MM (2011). Breakthroughs in the genetics of orofacial clefting. Trends in Molecular Medicine, 17, 725–733. [DOI] [PubMed] [Google Scholar]
- Marazita ML (2012). The evolution of human genetic studies of cleft lip and cleft palate. Annual Review of Genomics and Human Genetics, 13, 263–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Abadias N, Holmes G, Pankratz T, Wang Y, Zhou X, Jabs EW, et al. (2013). From shape to cells: Mouse models reveal mechanisms altering palate development in Apert syndrome. Disease Models & Mechanisms, 6, 768–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo R, Freer AM, Zinyk DL, Crackower MA, Michaud J, Heng HH, et al. (1997). Specific and redundant functions of Gli2 and Gli3 zinc finger genes in skeletal patterning and development. Development, 124, 113–123. [DOI] [PubMed] [Google Scholar]
- Moretti F, Marinari B, Lo Iacono N, Botti E, Giunta A, Spallone G, et al. (2010). A regulatory feedback loop involving p63 and IRF6 links the pathogenesis of 2 genetically different human ectodermal dysplasias. The Journal of Clinical Investigation, 120, 1570–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muenke M, Schell U, Hehr A, Robin NH, Losken HW, Schinzel A, et al. (1994). A common mutation in the fibroblast growth factor receptor 1 gene in Pfeiffer syndrome. Nature Genetics, 8, 269–274. [DOI] [PubMed] [Google Scholar]
- Muraoka N, Shum L, Fukumoto S, Nomura T, Ohishi M, & Nonaka K (2005). Transforming growth factor-beta3 promotes mesenchymal cell proliferation and angiogenesis mediated by the enhancement of cyclin D1, Flk-1, and CD31 gene expression during CL/Fr mouse lip fusion. Birth Defects Research. Part A, Clinical and Molecular Teratology, 73, 956–965. [DOI] [PubMed] [Google Scholar]
- Mustonen T, Pispa J, Mikkola ML, Pummila M, Kangas AT, Pakkasjarvi L, et al. (2003). Stimulation of ectodermal organ development by Ectodysplasin-A1. Developmental Biology, 259, 123–136. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Colbert M, Krenz M, Molkentin JD, Hahn HS, Dorn GW 2nd., et al. (2007). Mediating ERK 1/2 signaling rescues congenital heart defects in a mouse model of Noonan syndrome. The Journal of Clinical Investigation, 117, 2123–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatomi M, Wang XP, Key D, Lund JJ, Turbe-Doan A, Kist R, et al. (2010). Genetic interactions between Pax9 and Msx1 regulate lip development and several stages of tooth morphogenesis. Developmental Biology, 340, 438–449. [DOI] [PubMed] [Google Scholar]
- Nelson DK, & Williams T (2004). Frontonasal process-specific disruption of AP-2alpha results in postnatal midfacial hypoplasia, vascular anomalies, and nasal cavity defects. Developmental Biology, 267, 72–92. [DOI] [PubMed] [Google Scholar]
- Niihori T, Aoki Y, Narumi Y, Neri G, Cave H, Verloes A, et al. (2006). Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome. Nature Genetics, 38, 294–296. [DOI] [PubMed] [Google Scholar]
- Noonan JA (1968). Hypertelorism with Turner phenotype. A new syndrome with associated congenital heart disease. American Journal of Diseases of Children, 116, 373–380. [DOI] [PubMed] [Google Scholar]
- Nottoli T, Hagopian-Donaldson S, Zhang J, Perkins A, & Williams T (1998). AP-2-null cells disrupt morphogenesis of the eye, face, and limbs in chimeric mice. Proceedings of the National Academy of Sciences of the United States of America, 95, 13714–13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldridge M, Zackai EH, McDonald-McGinn DM, Iseki S, Morriss-Kay GM, Twigg SR, et al. (1999). De novo alu-element insertions in FGFR2 identify a distinct pathological basis for Apert syndrome. American Journal of Human Genetics, 64, 446–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchal J, & Uttchin V (2003). Management of craniosynostosis. Plastic and Reconstructive Surgery, 111, 2032–2048. quiz 2049. [DOI] [PubMed] [Google Scholar]
- Pandit B, Sarkozy A, Pennacchio LA, Carta C, Oishi K, Martinelli S, et al. (2007). Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nature Genetics, 39, 1007–1012. [DOI] [PubMed] [Google Scholar]
- Parada C, & Chai Y (2012). Roles of BMP signaling pathway in lip and palate development. Frontiers of Oral Biology, 16, 60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park WJ, Theda C, Maestri NE, Meyers GA, Fryburg JS, Dufresne C, et al. (1995). Analysis of phenotypic features and FGFR2 mutations in Apert syndrome. American Journal of Human Genetics, 57, 321–328. [PMC free article] [PubMed] [Google Scholar]
- Pauws E, Hoshino A, Bentley L, Prajapati S, Keller C, Hammond P, et al. (2009). Tbx22null mice have a submucous cleft palate due to reduced palatal bone formation and also display ankyloglossia and choanal atresia phenotypes. Human Molecular Genetics, 18, 4171–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlyn CA, Morriss-Kay G, Darvann T, Tenenbaum M, & Ornitz DM (2006). A model for the pharmacological treatment of crouzon syndrome. Neurosurgery, 59, 210–215. discussion 210–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer RA (1964). Dominant hereditary acrocephalosyndactylia. Zeitschrift für Kinderheilkunde, 90, 301–320. [PubMed] [Google Scholar]
- Phelps PD, Poswillo D, & Lloyd GA (1981). The ear deformities in mandibulofacial dysostosis (Treacher Collins syndrome). Clinical Otolaryngology and Allied Sciences, 6, 15–28. [DOI] [PubMed] [Google Scholar]
- Pierpont ME, Magoulas PL, Adi S, Kavamura MI, Neri G, Noonan J, et al. (2014). Cardio-facio-cutaneous syndrome: Clinical features, diagnosis, and management guidelines. Pediatrics, 134, e1149–e1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro M, & Freire-Maia N (1994). Ectodermal dysplasias: A clinical classification and a causal review. American Journal of Medical Genetics, 53, 153–162. [DOI] [PubMed] [Google Scholar]
- Pispa J, Jung HS, Jernvall J, Kettunen P, Mustonen T, Tabata MJ, et al. (1999). Cusp patterning defect in Tabby mouse teeth and its partial rescue by FGF. Developmental Biology, 216, 521–534. [DOI] [PubMed] [Google Scholar]
- Proetzel G, Pawlowski SA, Wiles MV, Yin M, Boivin GP, Howles PN, et al. (1995). Transforming growth factor-beta 3 is required for secondary palate fusion. Nature Genetics, 11, 409–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauen KA (2007). HRAS and the Costello syndrome. Clinical Genetics, 71, 101–108. [DOI] [PubMed] [Google Scholar]
- Rauen KA, Banerjee A, Bishop WR, Lauchle JO, McCormick F, McMahon M, et al. (2011). Costello and cardio-facio-cutaneous syndromes: Moving toward clinical trials in RASopathies. American Journal of Medical Genetics. Part C, Seminars in Medical Genetics, 157C, 136–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauen KA, Huson SM, Burkitt-Wright E, Evans DG, Farschtschi S, Ferner RE, et al. (2015). Recent developments in neurofibromatoses and RASopathies: Management, diagnosis and current and future therapeutic avenues. American Journal of Medical Genetics Part A, 167A, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzaque MA, Nishizawa T, Komoike Y, Yagi H, Furutani M, Amo R, et al. (2007). Germline gain-of-function mutations in RAF1 cause Noonan syndrome. Nature Genetics, 39, 1013–1017. [DOI] [PubMed] [Google Scholar]
- Reardon W, Winter RM, Rutland P, Pulleyn LJ, Jones BM, & Malcolm S (1994). Mutations in the fibroblast growth factor receptor 2 gene cause Crouzon syndrome. Nature Genetics, 8, 98–103. [DOI] [PubMed] [Google Scholar]
- Rice R, Spencer-Dene B, Connor EC, Gritli-Linde A, McMahon AP, Dickson C, et al. (2004). Disruption of Fgf10/Fgfr2b-coordinated epithelial-mesenchymal interacttions causes cleft palate. The Journal of Clinical Investigation, 113, 1692–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson RJ, Dixon J, Malhotra S, Hardman MJ, Knowles L, Boot-Handford RP, et al. (2006). Irf6 is a key determinant of the keratinocyte proliferation-differentiation switch. Nature Genetics, 38, 1329–1334. [DOI] [PubMed] [Google Scholar]
- Roberts AE, Araki T, Swanson KD, Montgomery KT, Schiripo TA,Joshi VA, et al. (2007). Germline gain-of-function mutations in SOS1 cause Noonan syndrome. Nature Genetics, 39, 70–74. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Tetsu O, Tidyman WE, Estep AL, Conger BA, Cruz MS, et al. (2006). Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science, 311, 1287–1290. [DOI] [PubMed] [Google Scholar]
- Rovin S, Dachi SF, Borenstein DB, & Cotter WB (1964). Mandibulofacial dysostosis, a familial study of five generations. The Journal of Pediatrics, 65, 215–221. [DOI] [PubMed] [Google Scholar]
- Sadier A, Viriot L, Pantalacci S, & Laudet V (2014). The ectodysplasin pathway: From diseases to adaptations. Trends in Genetics, 30(1), 24–31. [DOI] [PubMed] [Google Scholar]
- Salisbury DM, & Stothers JK (1981). Hypohidrotic ectodermal dysplasia and sudden infant death. Lancet, 1, 153–154. [DOI] [PubMed] [Google Scholar]
- Satokata I, & Maas R (1994). Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nature Genetics, 6, 348–356. [DOI] [PubMed] [Google Scholar]
- Schell U, Hehr A, Feldman GJ, Robin NH, Zackai EH, de Die-Smulders C, et al. (1995). Mutations in FGFR1 and FGFR2 cause familial and sporadic Pfeiffer syndrome. Human Molecular Genetics, 4, 323–328. [DOI] [PubMed] [Google Scholar]
- Schubbert S, Bollag G, & Shannon K (2007). Deregulated Ras signaling in developmenttal disorders: New tricks for an old dog. Current Opinion in Genetics & Development, 17, 15–22. [DOI] [PubMed] [Google Scholar]
- Schubbert S, Zenker M, Rowe SL, Boll S, Klein C, Bollag G, et al. (2006). Germline KRAS mutations cause Noonan syndrome. Nature Genetics, 38, 331–336. [DOI] [PubMed] [Google Scholar]
- Schuffenhauer S, Leifheit HJ, Lichtner P, Peters H, Murken J, & Emmerich P (1999). De novo deletion (14)(q11.2q13) including PAX9: Clinical and molecular findings. Journal of Medical Genetics, 36, 233–236. [PMC free article] [PubMed] [Google Scholar]
- Schuhmacher AJ, Guerra C, Sauzeau V, Canamero M, Bustelo XR, &Barbacid M (2008). A mouse model for Costello syndrome reveals an Ang II-mediated hypertensive condition. The Journal of Clinical Investigation, 118, 2169–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seegmiller R, Fraser FC, & Sheldon H (1971). A new chondrodystrophic mutant in mice. Electron microscopy of normal and abnormal chondrogenesis. The Journal of Cell Biology, 48, 580–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla V, Coumoul X, Wang RH, Kim HS, & Deng CX (2007). RNA interference and inhibition of MEK-ERK signaling prevent abnormal skeletal phenotypes in a mouse model of craniosynostosis. Nature Genetics, 39, 1145–1150. [DOI] [PubMed] [Google Scholar]
- Slaney SF, Oldridge M, Hurst JA, Moriss-Kay GM, Hall CM, Poole MD, et al. (1996). Differential effects of FGFR2 mutations on syndactyly and cleft palate in Apert syndrome. American Journal of Human Genetics, 58, 923–932. [PMC free article] [PubMed] [Google Scholar]
- Smith J (1929). Hereditary ectodermal dysplasia. Archives of Disease in Childhood, 4, 215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon LM, & Keuer EJ (1980). The ectodermal dysplasias. Problems of classification and some newer syndromes. Archives of Dermatology, 116, 1295–1299. [DOI] [PubMed] [Google Scholar]
- Sperry ED, Hurd EA, Durham MA, Reamer EN, Stein AB, & Martin DM (2014). The chromatin remodeling protein CHD7, mutated in CHARGE syndrome, is necessary for proper craniofacial and tracheal development. Developmental Dynamics: An Official Publication of the American Association of Anatomists, 243, 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava AK, Durmowicz MC, Hartung AJ, Hudson J, Ouzts LV, Donovan DM, et al. (2001). Ectodysplasin-A1 is sufficient to rescue both hair growth and sweat glands in Tabby mice. Human Molecular Genetics, 10, 2973–2981. [DOI] [PubMed] [Google Scholar]
- Srivastava AK, Pispa J, Hartung AJ,Du Y, Ezer S,Jenks T,et al. (1997). The Tabby phenotype is caused by mutation in a mouse homologue of the EDA gene that reveals novel mouse and human exons and encodes a protein (ectodysplasin-A) with collagenous domains. Proceedings of the National Academy of Sciences of the United States of America, 94, 13069–13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stottmann RW, Bjork BC, Doyle JB, & Beier DR (2010). Identification of a Van der Woude syndrome mutation in the cleft palate 1 mutant mouse. Genesis, 48, 303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia M, Mehler EL, Goldberg R, Zampino G, Brunner HG, Kremer H, et al. (2001). Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nature Genetics, 29, 465–468. [DOI] [PubMed] [Google Scholar]
- Tartaglia M, Pennacchio LA, Zhao C, Yadav KK, Fodale V, Sarkozy A, et al. (2007). Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nature Genetics, 39, 75–79. [DOI] [PubMed] [Google Scholar]
- Taya Y, O’Kane S, & Ferguson MW (1999). Pathogenesis of cleft palate in TGF-beta3 knockout mice. Development, 126, 3869–3879. [DOI] [PubMed] [Google Scholar]
- Thomason HA, Dixon MJ, & Dixon J (2008). Facial clefting in Tp63 deficient mice results from altered Bmp4, Fgf8 and Shh signaling. Developmental Biology, 321, 273–282. [DOI] [PubMed] [Google Scholar]
- Thurnam J (1848). Two cases in which the skin, hair and teeth were very imperfectly developed. Medico-Chirurgical Transactions, 31, 71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidyman WE, & Rauen KA (2009). The RASopathies: Developmental syndromes of Ras/MAPK pathway dysregulation. Current Opinion in Genetics & Development, 19, 230–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trokovic N, Trokovic R, Mai P, & Partanen J (2003). Fgfr1 regulates patterning of the pharyngeal region. Genes & Development, 17, 141–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urosevic J, Sauzeau V, Soto-Montenegro ML, Reig S, Desco M, Wright EM, et al. (2011). Constitutive activation of B-Raf in the mouse germ line provides a model for human cardio-facio-cutaneous syndrome. Proceedings of the National Academy of Sciences of the United States of America, 108, 5015–5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uslu VV, Petretich M, Ruf S, Langenfeld K, Fonseca NA, Marioni JC, et al. (2014). Long-range enhancers regulating Myc expression are required for normal facial morphogenesis. Nature Genetics, 46, 753–758. [DOI] [PubMed] [Google Scholar]
- Van Laer L, Dietz H, & Loeys B (2014). Loeys-Dietz syndrome. Advances in Experimental Medicine and Biology, 802, 95–105. [DOI] [PubMed] [Google Scholar]
- Vogels A, & Fryns JP (2006). Pfeiffer syndrome. Orphanet Journal of Rare Diseases, 1, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, et al. (2004). Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell, 116, 855–867. [DOI] [PubMed] [Google Scholar]
- Wang C, Chang JY, Yang C, Huang Y, Liu J, You P,et al. (2013). Type 1 fibroblast growth factor receptor in cranial neural crest cell-derived mesenchyme is required for palatogenesis. The Journal of Biological Chemistry, 288, 22174–22183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sun M, Uhlhorn VL, Zhou X, Peter I, Martinez-Abadias N, et al. (2010). Activation of p38 MAPK pathway in the skull abnormalities of Apert syndrome Fgfr2 (+P253R) mice. BMC Developmental Biology, 10, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xiao R, Yang F, Karim BO, Iacovelli AJ, Cai J, et al. (2005). Abnormalities in cartilage and bone development in the Apert syndrome FGFR2(+/S252W) mouse. Development, 132, 3537–3548. [DOI] [PubMed] [Google Scholar]
- Wassif CA, Zhu P, Kratz L, Krakowiak PA, Battaile KP, Weight FF, et al. (2001). Biochemical, phenotypic and neurophysiological characterization of a genetic mouse model of RSH/Smith–Lemli–Opitz syndrome. Human Molecular Genetics, 10, 555–564. [DOI] [PubMed] [Google Scholar]
- Wilkie AO, Oldridge M, Tang Z, & Maxson RE Jr. (2001). Craniosynostosis and related limb anomalies. Novartis Foundation Symposium, 232, 122–133. discussion 133–143. [DOI] [PubMed] [Google Scholar]
- Wilkie AO, Slaney SF, Oldridge M, Poole MD, Ashworth GJ, Hockley AD, et al. (1995). Apert syndrome results from localized mutations of FGFR2 and is allelic with Crouzon syndrome. Nature Genetics, 9, 165–172. [DOI] [PubMed] [Google Scholar]
- Winnier GE, Hargett L, & Hogan BL (1997). The winged helix transcription factor MFH1 is required for proliferation and patterning of paraxial mesoderm in the mouse embryo. Genes & Development, 11, 926–940. [DOI] [PubMed] [Google Scholar]
- Wu X, Simpson J, Hong JH, Kim KH, Thavarajah NK, Backx PH, et al. (2011). MEK-ERK pathway modulation ameliorates disease phenotypes in a mouse model of Noonan syndrome associated with the Raf1(L613V) mutation. The Journal of Clinical Investigation, 121, 1009–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Han J, Ito Y, Bringas P Jr., Urata MM, & Chai Y (2006). Cell autonomous requirement for Tgfbr2 in the disappearance of medial edge epithelium during palatal fusion. Developmental Biology, 297, 238–248. [DOI] [PubMed] [Google Scholar]
- Yin L, Du X, Li C, Xu X, Chen Z, Su N, et al. (2008). A Pro253Arg mutation in fibroblast growth factor receptor 2 (Fgfr2) causes skeleton malformation mimicking human Apert syndrome by affecting both chondrogenesis and osteogenesis. Bone, 42, 631–643. [DOI] [PubMed] [Google Scholar]
- Yu K, Herr AB, Waksman G, & Ornitz DM (2000). Loss of fibroblast growth factor receptor 2 ligand-binding specificity in Apert syndrome. Proceedings of the National Academy of Sciences of the United States of America, 97, 14536–14541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, & Ornitz DM (2001). Uncoupling fibroblast growth factor receptor 2 ligand binding specificity leads to Apert syndrome-like phenotypes. Proceedings of the National Academy of Sciences of the United States of America, 98, 3641–3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaritsky JJ, Eckman DM, Wellman GC, Nelson MT, & Schwarz TL (2000). Targeted disruption of Kir2.1 and Kir2.2 genes reveals the essential role of the inwardly rectifying K(+) current in K(+)-mediated vasodilation. Circulation Research, 87, 160–166. [DOI] [PubMed] [Google Scholar]
- Zenker M (2009). Genetic and pathogenetic aspects of Noonan syndrome and related disorders. Hormone Research, 72(Suppl. 2), 57–63. [DOI] [PubMed] [Google Scholar]
- Zenker M, Lehmann K, Schulz AL, Barth H, Hansmann D, Koenig R, et al. (2007). Expansion of the genotypic and phenotypic spectrum in patients with KRAS germline mutations. Journal of Medical Genetics, 44, 131–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Blackwell EL, McKnight MT, Knutsen GR, Vu WT, & Ruest LB (2012). Specific inactivation of Twist1 in the mandibular arch neural crest cells affects the development of the ramus and reveals interactions with hand2. Developmental Dynamics: An Official Publication of the American Association of Anatomists, 241, 924–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Song Y, Zhao X, Zhang X, Fermin C, & Chen Y (2002). Rescue of cleft palate in Msx1-deficient mice by transgenic Bmp4 reveals a network of BMP and Shh signaling in the regulation of mammalian palatogenesis. Development, 129, 4135–4146. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Li MJ, Zhu WL, Guo JZ, Wang Y, Li Y, et al. (2013). Association between interferon regulatory factor 6 gene polymorphisms and nonsyndromic cleft lip with or without cleft palate in a Chinese population. The Cleft Palate-Craniofacial Journal, 50, 570–576. [DOI] [PubMed] [Google Scholar]
- Zhou YX, Xu X, Chen L, Li C, Brodie SG, & Deng CX (2000). A Pro250Arg substitution in mouse Fgfr1 causes increased expression of Cbfa1 andpremature fusion of calvarial sutures. Human Molecular Genetics, 9, 2001–2008. [DOI] [PubMed] [Google Scholar]
- Zonana J, Elder ME, Schneider LC, Orlow SJ, Moss C, Golabi M, et al. (2000). A novel X-linked disorder of immune deficiency and hypohidrotic ectodermal dysplasia is allelic to incontinentia pigmenti and due to mutations in IKK-gamma (NEMO). American Journal of Human Genetics, 67, 1555–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchero TM, Cooper ME, Maher BS, Daack-Hirsch S, Nepomuceno B, Ribeiro L, et al. (2004). Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. The New England Journal of Medicine, 351, 769–780. [DOI] [PubMed] [Google Scholar]