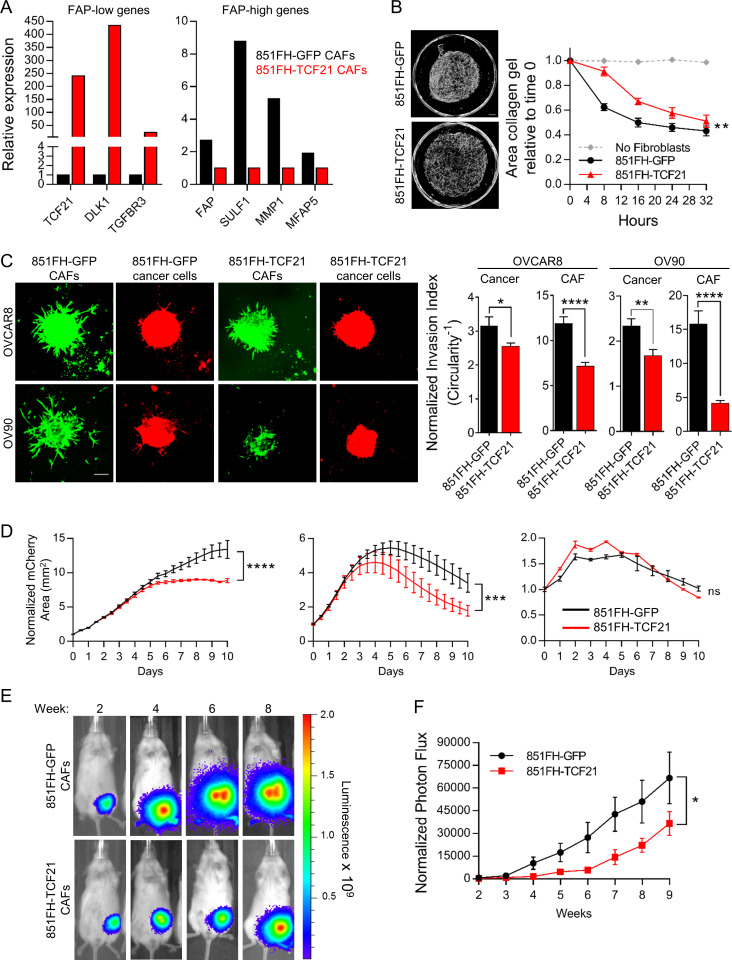
Figure 7.
Overexpression of TCF21 in FAP-high CAFs inhibits their protumorigenic functions. (A) qRT-PCR for FAP-low genes (TCF21, DLK1, and TGFBR3) and FAP-high genes (FAP, SULF1, MMP1, and MFAP5) in control versus TCF21 overexpressing 851FH CAFs. Fold-differences in gene expression were determined using the ΔΔ−Ct method, using UBC as a housekeeping gene, with three technical replicates per sample assayed. (B) Representative images of collagen gels cultured for 32 h with 851FH-GFP CAFs or 851-TCF21 CAFs (left); quantification of gel contraction over time (right). Data are presented as mean ± SEM, n = 3. **, P < 0.01, two-way ANOVA. Scale bar = 500 µm. (C) Representative images of spheroids generated using mCherry-expressing HGSOC cell lines OVCAR8 or OV90 with 851FH-GFP or 851FH-TCF21 CAFs mixed in. Spheroids were embedded in Matrigel and imaged after 4 d (left). Circularity was quantified in both channels (green, CAFs; red, cancer cells) using ImageJ (right). Data are presented as mean ± SEM, n = 10 spheroids per condition. *, P < 0.05; **, P < 0.01; ****, P < 0.0001, Student’s t test. Scale bar = 100 µm. (D) Growth curves of mCherry-labeled cancer cells co-cultured with 851FH-GFP or 851FH-TCF21 CAFs after treatment with 10 µM carboplatin. Data were normalized to the mCherry+ area at day 0 and are presented as mean ± SEM, n = 3. Asterisks reflect differences at the end of 10 d. ***, P < 0.001; ****, P < 0.0001, Student’s t test. (E) Spheroids of luciferase-expressing OVCAR8 cells alone or with 851FH-GFP or 851FH-TCF21 CAFs mixed in were generated as for invasion assays. After 72 h of spheroid formation, individual spheroids were implanted into the mammary fat pads of NSG mice. Representative images of individual mice are shown. (F) Quantification of luciferase signal, normalized to an unimplanted mouse over a period of 9 wk. Data are presented as mean ± SEM, n = 10–15 (animals in which tumors failed to grow were not included). *, P < 0.05, two-way ANOVA. FH, FAP high; FL, FAP low; ns, not significant.