Fig. 1. Annotation of methylation variable regulatory elements in developing mouse tissues.
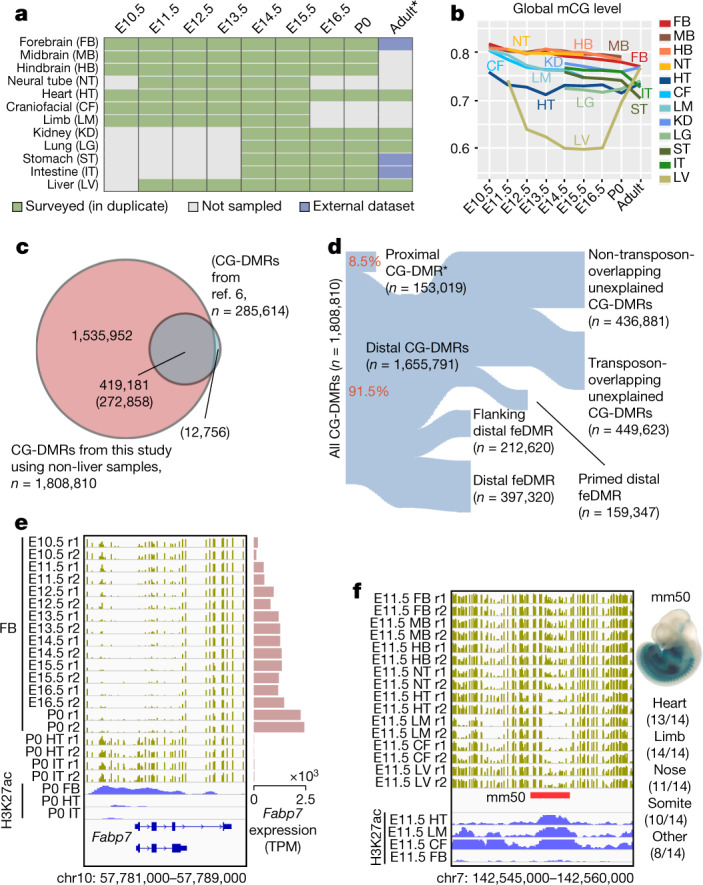
a, Tissue samples (green) profiled in this study. Blue cells indicate published data, grey cells indicate tissues and stages that were not sampled because either the organ is not yet formed or it was not possible to obtain sufficient material for the experiment, or the tissue was too heterogeneous to obtain informative data. *Additional data were generated in duplicate for adult tissues. b, Global mCG level of each tissue across their developmental trajectories. The adult forebrain was approximated using postnatal six-week-old frontal cortex9. c, Fetal CG-DMRs identified in this study encompass the majority of the adult CG-DMRs from a previous study11. Numbers with and without parentheses are related to fetal CG-DMRs and adult CG-DMRs, respectively. d, Categorization of CG-DMRs. Proximal CG-DMRs are those that overlap with promoters, CGIs or CGI shores. The rest are distal CG-DMRs. Fetal enhancer-linked CG-DMRs (feDMRs) are those that are predicted to be putative enhancers; those within 1 kb of distal feDMRs are flanking distal feDMRs. The remaining distal CG-DMRs showing hypomethylation are primed distal feDMRs. The rest are unexplained distal CG-DMRs, the functions of which are unknown, and they are further stratified according to their overlap with transposons (Methods). *Proximal CG-DMRs include 70,821 proximal feDMRs. e, mCG, H3K27ac and expression dynamics of Fabp7. Gold ticks represent CG sites; height represents mCG level, ranging from 0 to 1. The bottom three tracks show input-normalized H3K27ac enrichment in reads per kilobase per million mapped reads (RPKM), ranging from 0 to 20. Fabp7 expression in transcripts per million mapped reads (TPM) is shown on the right. f, mCG and H3K27ac profiles near an experimentally validated enhancer from the VISTA enhancer data set28. The data on the right show the number of embryos in which the enhancer element (ID in VISTA: mm50) was active in a given tissue (out of a total n = 14 embryos). The image shows the tissue where the tested enhancer was active in one representative embryo. r1 and r2 denote first and second replicate, respectively.
