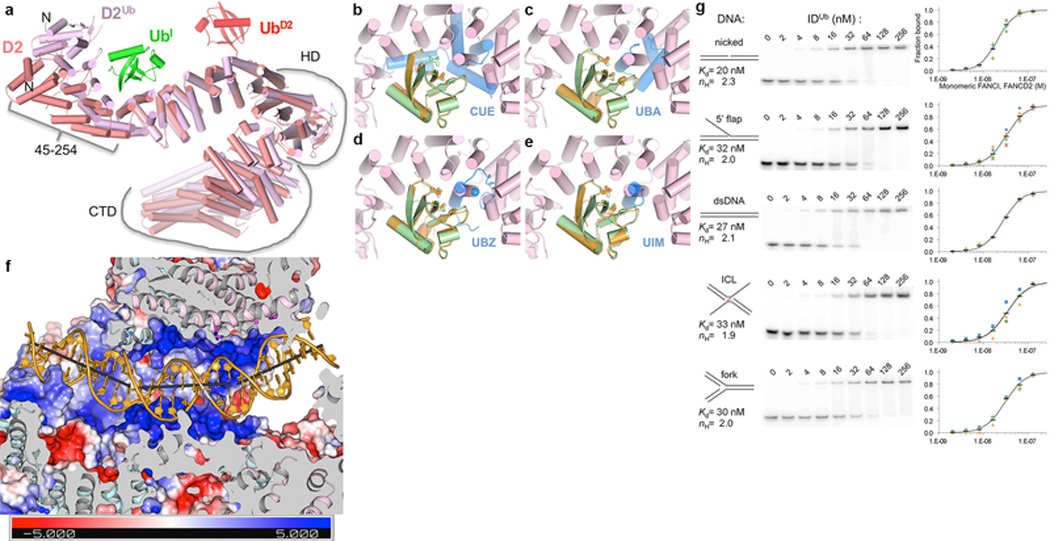
Extended Data Figure 9 |. Ubiquitination induces FANCD2 conformational changes associated with alternative FANCI-FANCD2 contacts; DNA binding activity of IDUb.
a, After ubiquitination, FANCI does not change significantly (entire molecule can be superimposed with a 1.6 Å r.m.s.d in 1132 Cα positions), but FANCD2 undergoes two changes. FANCD2 from the ID complex (salmon) is superimposed on that of IDUb (pink) by aligning the 2nd half of their NTDs (residues 255–587; 1.8 Å r.m.s.d. for 308 Cα positions), a segment that changes little on ubiquitination. UbD2 that is covalently attached to FANCD2 is in red, and the UbI with which it packs in green. The N-terminal portion of the NTD, which rotates by 38° towards FANCI as a mostly rigid body (residues 45 to 254, 0.68 Å r.m.s.d. for 202 Cα) is approximately marked by a bracket. This rotation at the center of the UbI binding site allows FANCD2 to better embrace UbI, and also to interact with FANCI (NTD-HD junction, residues 529 to 593), the latter involving similar residues on FANCD2 but mostly different ones on FANCI. Similarly marked is the HD domain (residues 604–928) that rotates relative to the NTD by 15°. Additional tilting of helices within the HD domain results in the CTD that follows (residues 929 to C-termini also marked) being rotated by 20° degrees relative to the invariant portion of the NTD. b-e, The FANCI and FANCD2 ubiquitin binding structural elements are distinct from commonly occurring ubiquitin-binding domains. Superposition of the UbI (green) bound to FANCD2Ub (pink) on the ubiquitin (orange) bound to the dimeric Vps29 CUE domain (blue) from PDBID 1P3Q in b, to the Cbl-b UBA domain (PDBID 2OOP) in c, to the UBZ domain of Faap20 (PDBID 3WWQ) in d, and to the UIM domain of Vps27 (PDBID 1Q0W) in e. The ubiquitin hydrophobic patch residues (Leu8, Ile44, and Val70) are shown in stick representation for both ubiquitin molecules in each figure. The blue sphere in d is the Zn atom of the UBZ domain. Orientation similar to that of Figure 4a. It has been suggested that FANCD2 shares sequence homology with the CUE domain34. While the 47-residue region of proposed homology (residues 191–237) partially overlaps the Ub-binding site, its structure is unrelated to the CUE domain, and Ub binding by FANCD2 is distinct. f, Molecular surface colored according to electrostatic potential calculated with PYMOL in the absence of DNA (colored −5 to +5 kT blue to red), in the same view as Figure 4d and with structural elements and surfaces above the DNA similarly clipped to reveal the DNA. As with the ID-ICL DNA complex, the FANCIUb groove partially encircles the DNA through basic and polar residues from α33b and α36b on one side of the groove and α40, α42 on the other. Near the middle of the DNA, however, the extension of α48 that results from coiling with FANCD2Ub α50 (Fig. 3e) gives rise to a new semicircular basic groove, between α46 and α48 on one side and the NTD α19b and α20 on the other, into which dsDNA binds (Fig. 4d). This is associated with one of the two DNA bends, which redirects the duplex away from clashing with the new position of FANCD2. Thereafter, the second bend occurs as the dsDNA is redirected by the FANCD2 CTD, which uses the localized patch of α48 and α50 to bind to the duplex analogously to the ID-ICL DNA complex. The side chains near the DNA, shown in Figure 4d as sticks, are from FANCIUb residues Lys291 (α15), Lys397 (α19b), Ser411 (α20), Lys793, Thr794 on α33b, Lys898 (α36b), Lys980 (α40), Lys1026 (α42), Lys1164 (α46), and Thr1238, Arg1242, Arg1245 and Lys1248 on the extended α48; and from FANCD2Ub residues His1288, His1292, and Arg1299 on α48, and Thr1351, Arg1352 and Gln1355 on α50. The sites of DNA bending, of 26° and 31°, are centered on the 11th and 21st base pairs from the FANCI end, respectively, with large roll values over three base-pair steps. g, The IDUb Kd values for dsDNA, nicked, 5’ flap, ICL and fork DNA vary by less than a factor of two, with dsDNA and nicked DNA exhibiting slightly tighter binding. EMSA of the equimolar mixture of the mono-ubiquitinated FANCIUb and FANCD2Ub, each at the indicated concentrations, binding to the 32P-labeled DNA substrates (0.5 nM) shown schematically. The plots with a logarithmic X-axis show fraction bound in at least three repetitions of each experiment (different color and shape markers) and their mean value (black dash). As with the non-ubiquitinated complex, the binding isotherms fit a Hill slope model best. A binding curve (black line) simulated with the indicated Kd and Hill coefficient (ηH) values is shown on each plot.