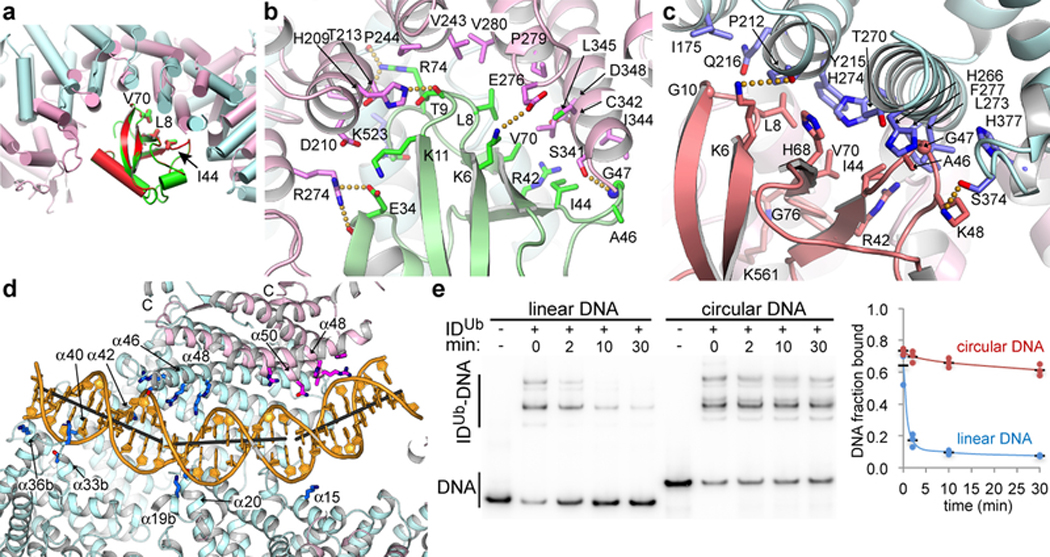
Figure 4 |. Interactions with ubiquitin and DNA.
a, Superposition of the FANCD2Ub-UbI and FANCIUb-UbD2 interfaces by aligning UbD2 (red) with UbI (green). The paralogs that each ubiquitin is attached to are not shown for clarity. The ubiquitin hydrophobic patch residues (Leu8, Ile44, and Val70) are shown as sticks for both ubiquitin molecules. Colored as in Fig. 2a. b, Close-up view of the non-covalent interface between FANCD2Ub (pink) and UbI (green) showing the residues within interaction distance as sticks (darker colors). Yellow dotted lines indicate potential hydrogen bonds. c, Close-up view of the reciprocal interface between FANCIUb (cyan) and UbD2 (green). d, Cartoon, colored as in Fig. 2a, showing IDUb side chains within contact distance of the DNA as sticks (darker hues). Gray sticks show the helical axes for the three relatively straight segments of the DNA. The majority of FANCD2Ub and parts of FANCIUb are clipped above the plane of the figure to make the DNA visible (Extended Data Fig. 9f legend lists side chains shown). e, Autoradiogram of DNA binding competition time course after 8 μM of unlabeled 67 bp dsDNA was added to the IDUb complex (800 nM) pre-assembled on a 95 bp circular nicked DNA or the corresponding linear nicked DNA, both at 400 nM with only 2 nM of each 32P labeled. 32P DNA-protein complexes contain multiple IDUb proteins due to the length of each DNA. Fraction of DNA bound is quantified in the chart (repetitions marked by circles, and their mean by a black dash; n=3).