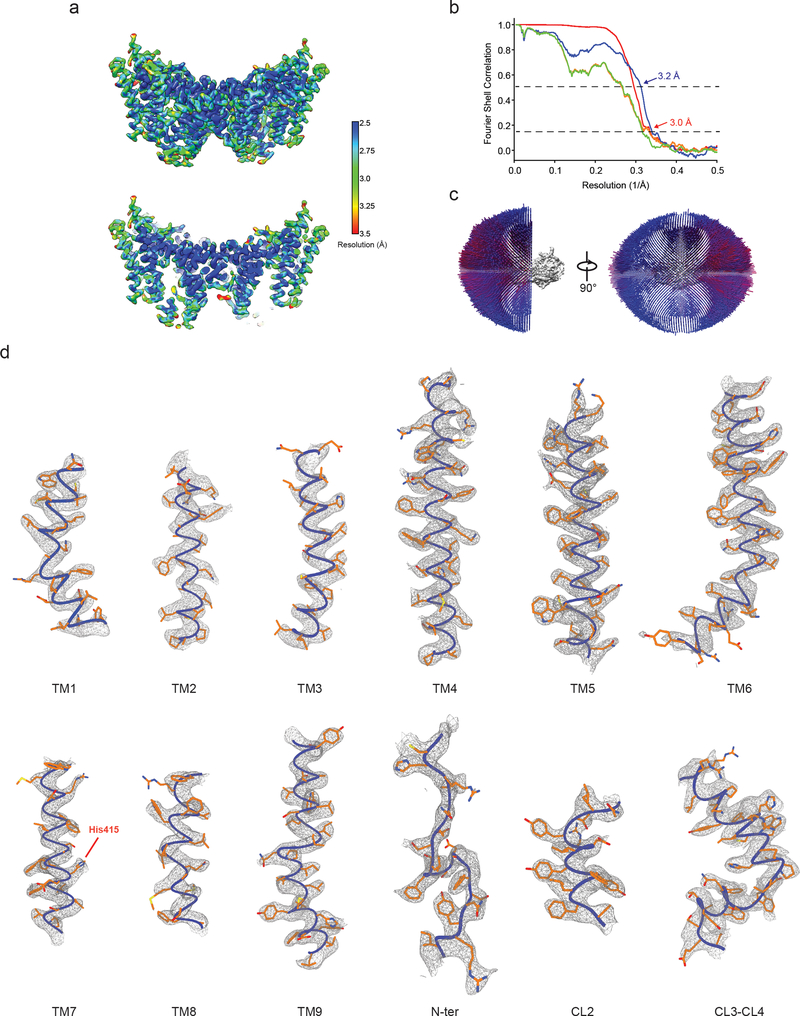
Extended Data Fig. 3 |. Single-particle cryo-EM analysis of DGAT1 reconstituted in PMAL-C8.
a, Local resolution of the final cryo-EM map of DGAT1. A sliced view of local resolution is shown in the lower panel. b, FSC curves: gold-standard FSC curve between the two half maps with indicated resolution at FSC = 0.143 (red); FSC curve between the atomic model and the final map with indicated resolution at FSC = 0.5 (blue); FSC curve between half map 1 (orange) or half map 2 (green) and the atomic model refined against half map 1. c, Cutaway views of angular distribution of particle images included in the final 3D reconstruction. d, Cryo-EM densities superimposed with the atomic model for individual transmembrane helices (TM1 – 9), resolved N-terminal region (N-ter), and helices in the cytosolic loop region (CL2 – 4). The conserved His415 is also labeled. Maps are contoured at 4 σ.