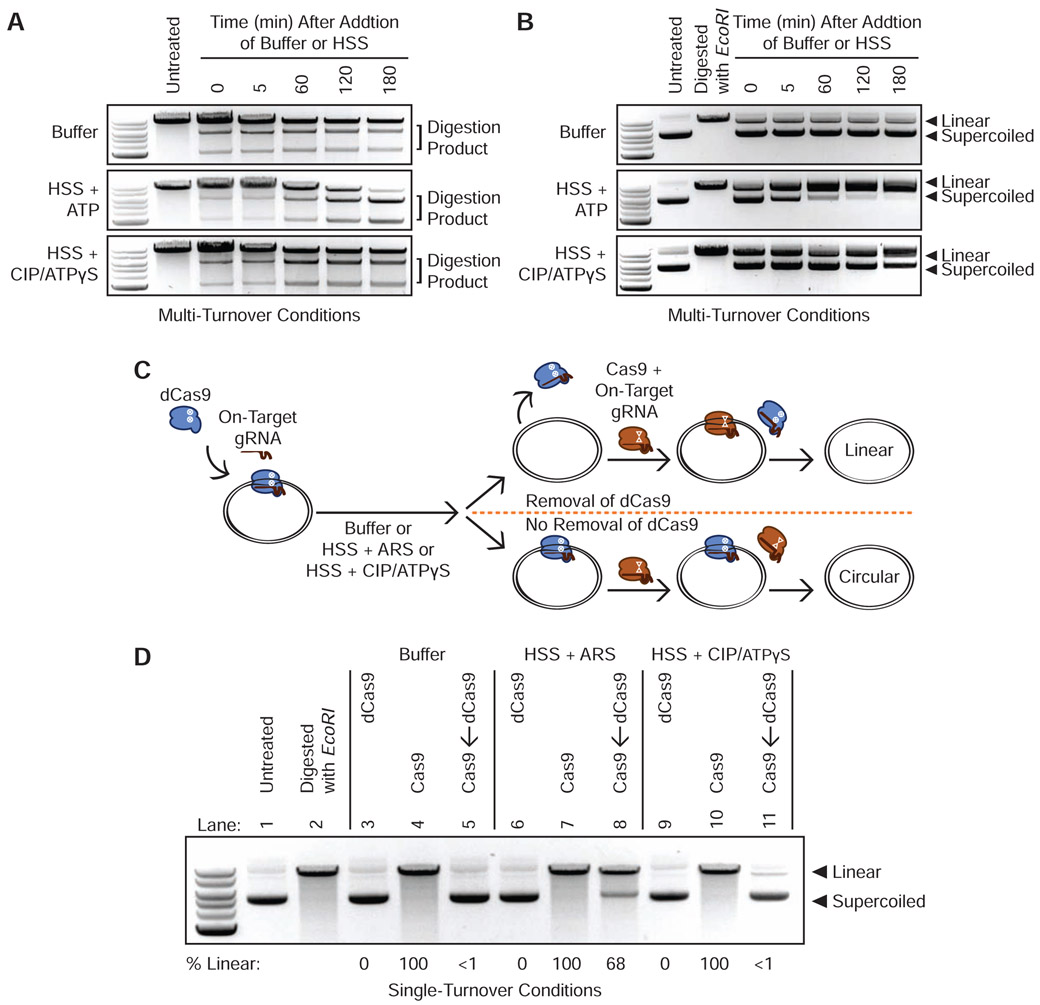
Figure 1. Energy-Dependent Release of Cas9 and dCas9 from DNA in HSS.
(A) Time course of Cas9 RNPs programmed against a linear DNA substrate in a 1:2 molar ratio in buffer, HSS with ATP, or HSS with CIP and ATPγS.
(B) Time course of Cas9 RNPs programmed against a plasmid substrate in a 1:2 molar ratio in buffer, HSS with ATP, or HSS with CIP and ATPγS.
(C) Schematic of the single-turnover competition assay.
(D) ATP-dependent unloading of dCas9 off a plasmid substrate in X. laevis HSS. Presence of linearized DNA after addition of a 10-fold molar excess of Cas9 indicates removal of dCas9 while persistence of circular DNA indicates stable binding of dCas9. See also Figure S1C.