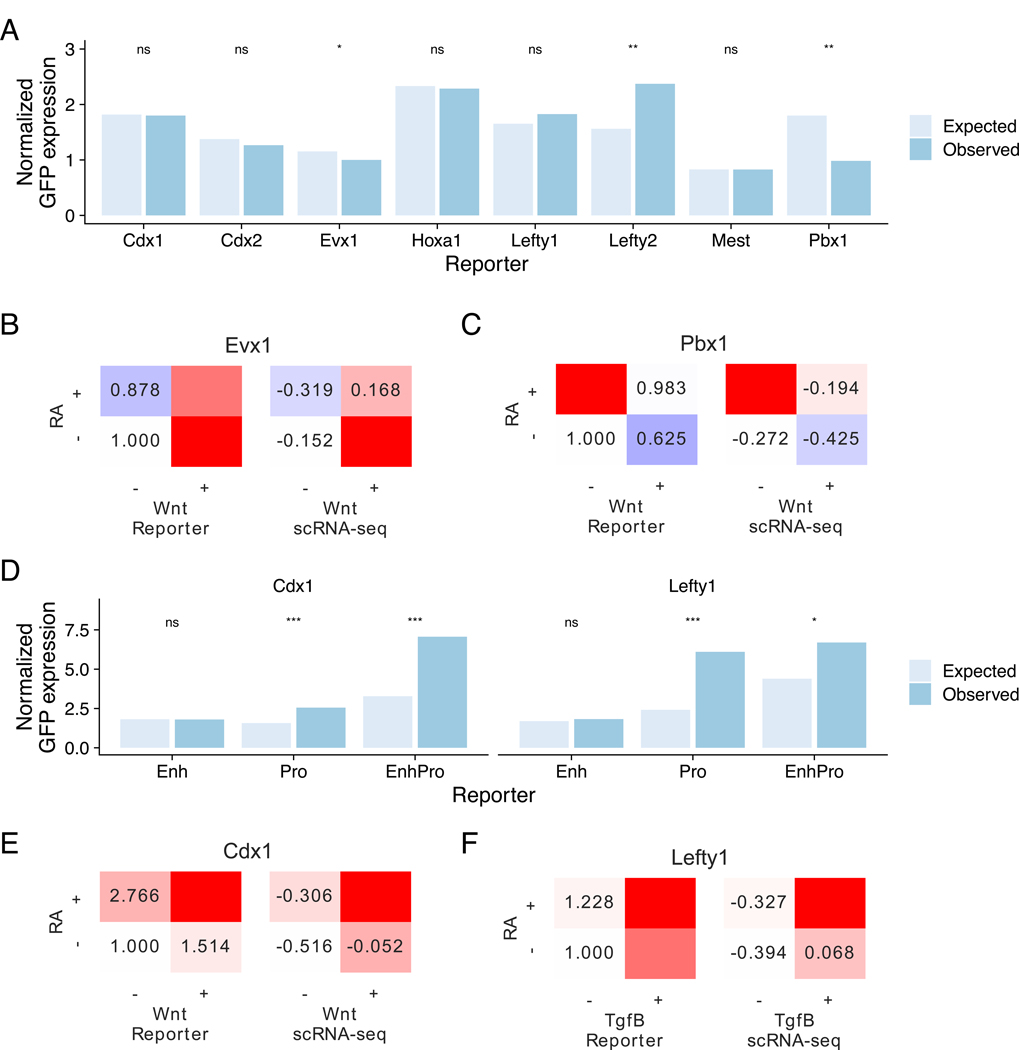
Figure 4. Sequence features implement signaling-based combinatorial control of gene expression.
(A) Expected and observed normalized GFP expression of enhancer reporter assays for genes chosen for validation. Expected expression was computed under a linear model where effects are assumed to be linearly additive. Asterisks denote result of paired t-tests on log expression. * is significant at p < 0.2, ** at p < 0.1.
(B, C) Mean expression of GFP reporters in enhancer reporter assay (left) and scRNA-seq (right) showing combinatorial effects. Expression in enhancer reporter assay is normalized to the −/− condition. (B) Assay results for Evx1, which is most highly expressed in RA-/Wnt+, (C) Assay results for Pbx1, which is most highly expressed in RA+/Wnt-
(D) Expected and observed normalized GFP expression of reporters containing sequence features from the enhancer, the native promoter, or both for Cdx1 and Lefty1. Expected expression was computed under a linear model where effects are assumed to be linearly additive. Asterisks denote result of paired t-tests. * is significant at p < 0.2, *** at p < 0.05.
(E, F) Mean expression of GFP reporters in enhancer reporter assay (left) and scRNA-seq (right) showing combinatorial effects. Expression in enhancer reporter assay is normalized to the −/− condition. (B) Assay results for Cdx1, which is most highly expressed in RA-/Wnt+, (C) Assay results for Lefty1, which is most highly expressed in RA+/Tgfβ+/Bmp- See also Figure S4