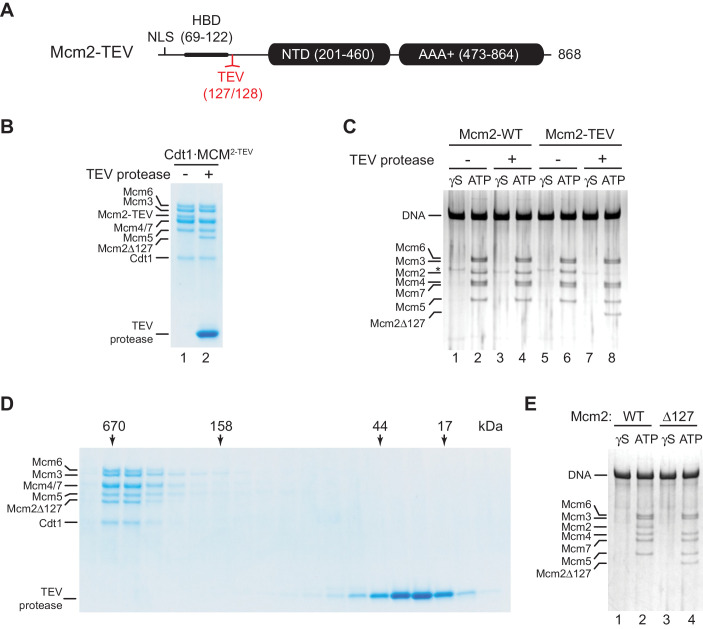
Figure 1. Residues 1–127 of the Mcm2 NTE are dispensable for MCM DH stability.
(A) Schematic of Mcm2 domain structure. Numbers indicate amino acid positions. The position of the TEV cleavage site is highlighted in red. NLS: Nuclear localization sequence; HBD: Histone binding domain; NTD: N-terminal domain; AAA+: ATPase domain. (B) Cdt1·MCM2-TEV was mock-treated or digested with TEV protease for 1 hr at 30°C, as indicated. Reactions were fractionated on SDS-PAGE and stained with Coomassie blue. (C) MCM loading reactions were performed on 3 kbp ARS305-containing DNA in the presence of ATPγS (γS) or ATP as indicated. DNA-bound material was washed with high-salt buffer, mock-treated or digested with TEV protease as indicated, washed again with high-salt buffer, and analyzed by SDS-PAGE and silver staining. * denotes Orc1 protein. (D) Gel-filtration analysis of purified Cdt1·MCM2-TEV following digestion with TEV protease. The digestion reaction was fractionated on a Superdex 200 column and fractions analyzed by SDS-PAGE and Coomassie stain. (E) Mcm2-7 loading reactions with either wildtype Cdt1·MCM (lanes 1+2) or Cdt1·MCM2-Δ127 (lanes 3+4). Reactions were performed either in the presence of ATPγS or ATP as indicated and DNA-beads subsequently washed with high-salt buffer. DNA-bound fractions were analyzed by SDS-PAGE and silver stain.
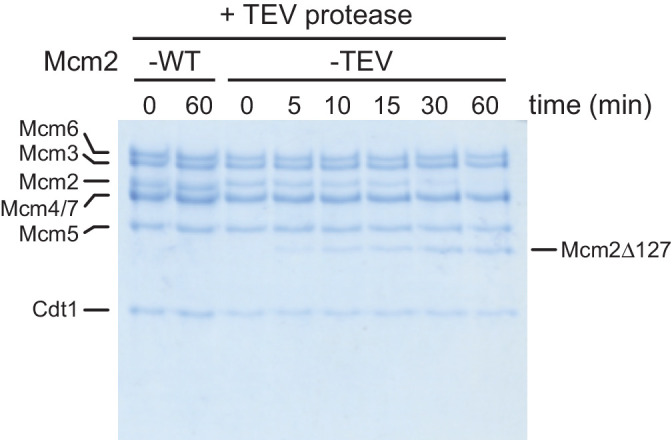
Figure 1—figure supplement 1. Time course analysis of Cdt1·MCM2-TEV and Cdt1·MCM2-WT cleavage by TEV protease.