Figure 1. Mutations in MSL3 cause a novel neurodevelopmental disorder.
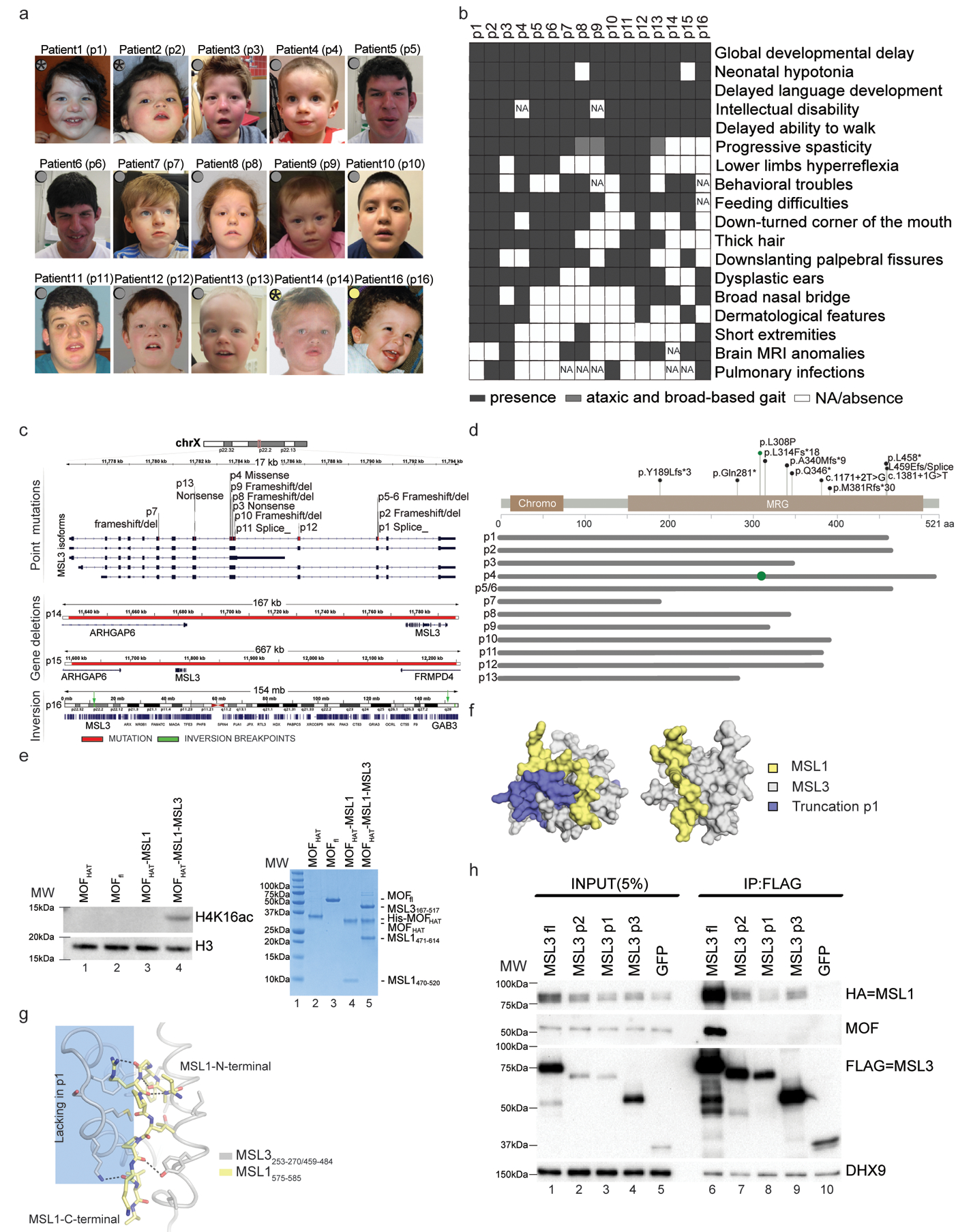
(a) Photographs of patients with MSL3 mutations. Gray circles indicate intragenic variants. Yellow circles indicate whole-gene deletions. Stars show the patients who donated skin biopsies. Written consent for the use of photographs was obtained from the parents of affected individuals. (b) Human Phenotype Ontology heat map of patients’ common clinical features. Blank boxes represent either absent or unreported symptoms. MRI, magnetic resonance imaging; NA, not analyzed. (c) The MSL3 locus and identified variants are highlighted in red. p1–13 variants occur within MSL3 (top). p13/p14 deletions span ARHGAP6, MSL3, and FRMD4 (middle). p16 chromosomal inversion and breakpoints (green) are between Xp22.2 and Xq28 (bottom). (d) Annotation of the pathogenic variants on the primary protein structure. Gray lines represent predicted proteins, where most SNVs (except for p.Leu308Pro; shown as green circle) result in MRG domain truncation. aa, amino acid. fs, frameshift. (e) Left, cropped H4K16ac and H3 immunoblot of a HAT assay performed on nucleosomal substrate with the indicated proteins (full-length (fl; 2–458) and HAT domain (174–458)). Right (lanes 2–5), Coomassie-stained gel of the assayed proteins. The experiment was repeated twice with similar results. MW, molecular weight (lane 1). (f) Modeled surface representation of the MSL1–MSL3 complex (Protein Data Bank: 2Y0N; MSL1, yellow; MSL3, gray). Missing parts in p1 are highlighted in purple (left) or removed (right). (g) Schematic representing the MSL1 (575–585, yellow) and MSL3 (253–270/459–484, gray) interaction interface. Hydrogen bonds are shown in black. α-helices 5/6, which are missing in p1, are shaded blue. (h) FLAG co-immunoprecipitations of FLAG-tagged full-length MSL3, MSL3 variants (p2, p1, and p3) and a GFP control with cropped immunoblots. MSL3-FLAG-tag, MSL3(p2,p1,p3)-FLAG-tag, GFP-FLAG-tag expression vectors were transiently co-transfected with MSL1-HA-tag expression vector into Flp-In T-Rex 293 cells. DHX9 serves as input loading control. The experiment was repeated 4 times with similar results.
