Figure 7.
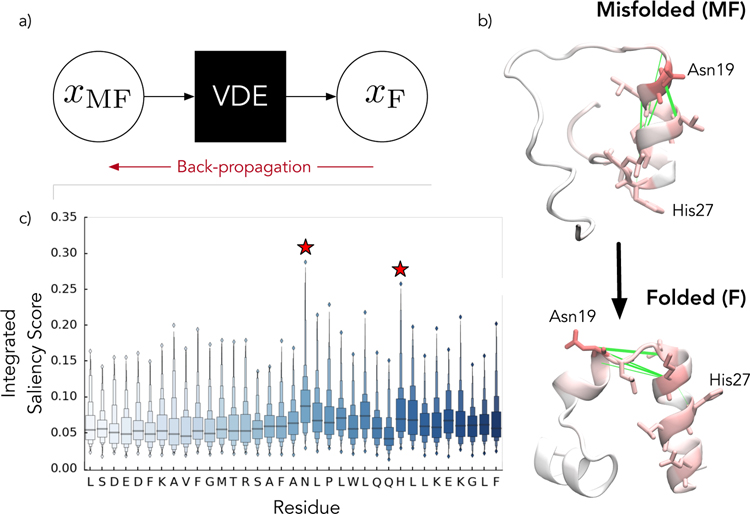
Protein saliency maps can be used to gain insight into the VDE. a) In saliency maps, the distances between the predicted and targeted output (i.e. contact distances in the folded states) are propagated back through the network to the input contact distances in order to gain insight into what the network learns. This is repeated for a large batch of possible configurations. b) For villin, the folded state is characterized by Cα contact distances to the central Asparagine residue. In the misfolded state, this residue is too close to the first helix forming non-native contacts. The green lines denote the 5 contacts with the highest median saliency scores. c) Integrating over the saliency at the atomic level allows us to infer the importances of individual residues in certain state transitions, making them prime candidates (red stars) for further biophysical characterization. The distributions are computed over 200 transitions.