Figure 1.
Capture of Molecules in Simulated Synapses
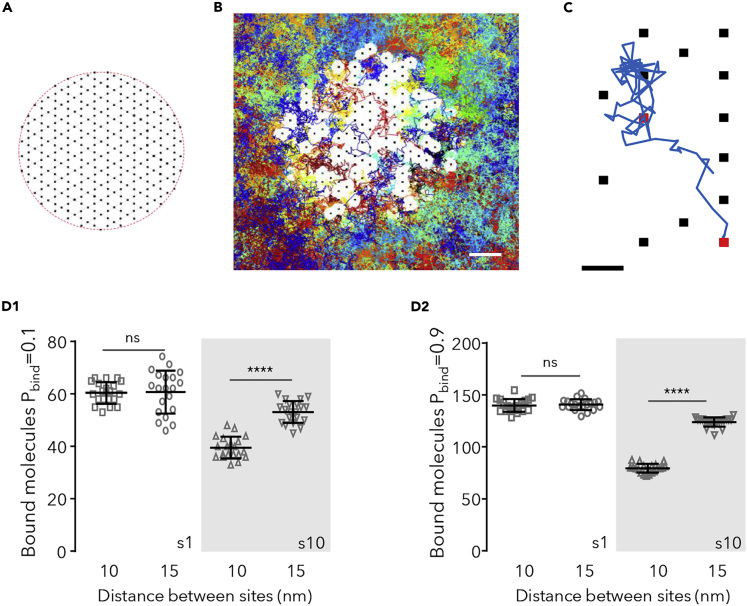
(A) Scheme of the hexagonal grid where binding sites could occur (black dots).
(B) Detail of the simulation space showing the randomly chosen binding sites (250 in total, black dots) overlaid with all the trajectories cumulated during 75s of simulation (200 molecules in total, 118 molecules bound at t = 75s, each trajectory depicted in a different color). The distance between sites was 15 nm. Molecules were 10 nm in diameter and a probability of binding (Pbind) of 0.9 and a probability of unbinding (Pfree) of 10−4. White spaces correspond to the excluded area due to the bound molecules (self-crowding). Bar: 50 nm.
(C) Detail of one trajectory (in blue) and binding sites (squares). The simulated molecule bound to the sites shown in red. Bar: 15 nm.
(D) Number of bound molecules when sites were distanced 10 nm or 15 nm, in steady state, for small (s1) or large (s10, gray area) molecules and low (D1) or high (D2) Pbind (values of 20 independent simulations overlaid with the mean ± SD, unpaired t test, ns: not significant; ∗∗∗∗p < 0.0001).