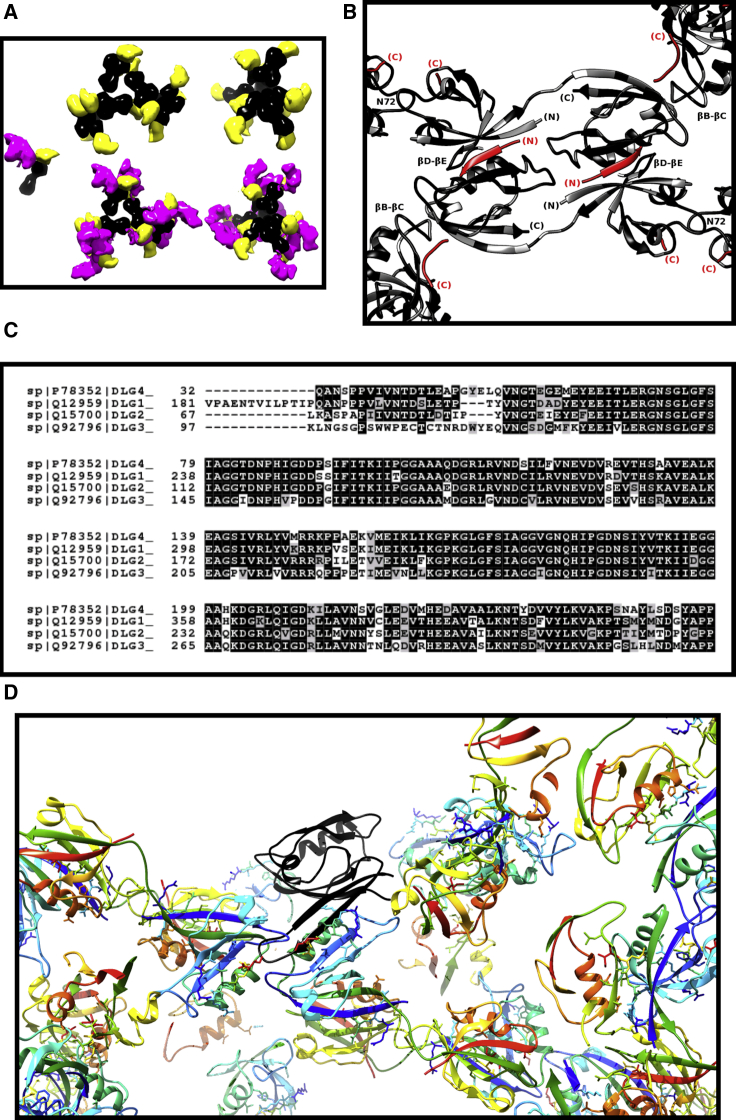
Figure 7.
Modulation of the PDZ1-2 scaffold. (A) shows views of a dodecamer (Fig. 5A, 12e) of PDZ1-2 drawn from the scaffolding space group decorated with PDZ3 and PDZ3-SH3GK. An isolated PDZ1-2-PDZ3-SH3GK model is shown leftmost. Molecules are represented in a similar way to Fig. 6, with PDZ1-2 in black, PDZ3 in yellow, and SH3GK in magenta. Sequence variation in the PDZ1-2 fragment across the MAGUK proteins (DLG1-4) is represented in (B), with the corresponding sequence alignment in (C). UniProt codes are shown alongside gene names in the alignment. The PDZ1-2 domains are shown in cartoon form, with the RRESEI ligand in red. The C- and N-termini of each PDZ1-2 are labeled in parentheses, the C-terminus of each RRESEI peptide bound at PDZ1 is labeled, and the N-terminus of each RRESEI peptide bound at PDZ2 is labeled. Selected structural elements at interfaces formed in the packing arrangement are also labeled. The PDZ1:PDZ1 interface is located by residue Asn 72 (N72); the αB(PDZ2)-βD-βE(PDZ1) interface is located by the βD-βE loop label and the αA(PDZ2)-βB-βC(PDZ1) interface by the βB-βC loop label. The sequence alignment obtained from the program ClustalW (72) is mapped onto the packing arrangement with the locations of identical residues colored black, similar residues colored gray, and residues with different properties colored white. (D) shows a section of the packing arrangement shown in Fig. 6A, with PDZ1-2 displayed in a cartoon form in a similar manner to Fig. 1; the nNOS PDZ domain (black) is shown added by docking on to the scaffold in association with PDZ1. Docking is accomplished via the hairpin loop of the nNOS PDZ domain inserting into the PDZ1 cleft (shown overlaying a bound RRESEI ligand in the figure). Molecular representations were generated with UCSF Chimera. To see this figure in color, go online.