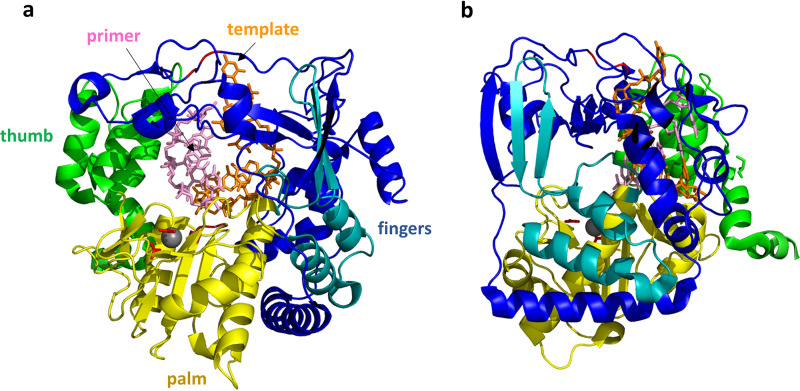
FIG 12.
FMDV 3Dpol/RNA elongation complex. (a) The view of 3Dpol is rotated 180° from the view shown in Fig. 9. The RNA template strand (orange) and primer strand (pink) are shown as sticks, while the protein is shown as a cartoon colored as described in the legend of Fig. 9. The 3Dpol structure largely resembles that of RV14 shown in Fig. 9. The FMDV 3Dpol structure was determined in the presence and absence of RNA, and little structural rearrangement was detected. The template RNA strand interacts mainly with the fingers region, while the primer strand interacts with the thumb and palm. A magnesium ion that binds near the catalytic site is shown as a gray sphere, with side chains of the coordinating D-238 and D-240 residues shown as red sticks. The positions of the two catalytic Asp residues, D-338 and D-339, are also indicated by the red loop to the right of the magnesium ion. (b) This view of the structure is rotated 90°. From this side view, it can be seen that the RNA double helix is positioned close to the right-hand surface, which is the top surface of the structure shown in panel a. During replication, new bases would be added to the 3′ end of the primer strand, extending that strand to the left. Thus, the double helix would need to proceed to the right (or, relatively speaking, the polymerase proceeds to the left) in order to processively position the new RNA 3′ end in the polymerase active site.