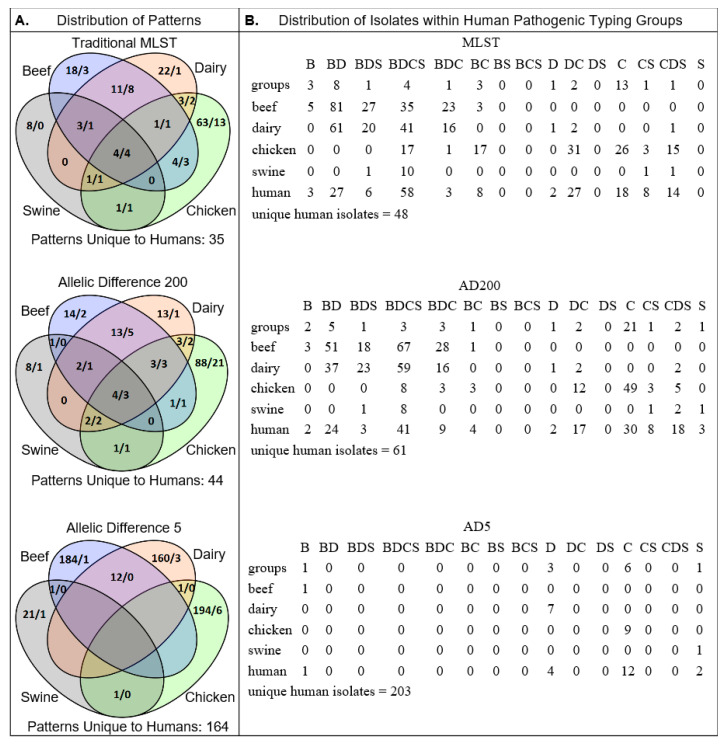
Figure 2.
(A) Venn diagrams showing the number of cgMLST patterns associated with each source for traditional MLST (upper), cgMLST200 (middle) and cgMLST5 (lower). Patterns are numbered by total (left) followed by those shared with human pathogenic strains (right). (B) Distribution of individual strains belonging to the human pathogenic patterns in A. Isolates recovered from humans found in typing groups unique to humans are listed below for each typing scheme. Group totals correspond with A. and source identification is as follows: Beef (B), Dairy (D), Swine (S) and Chicken (C).