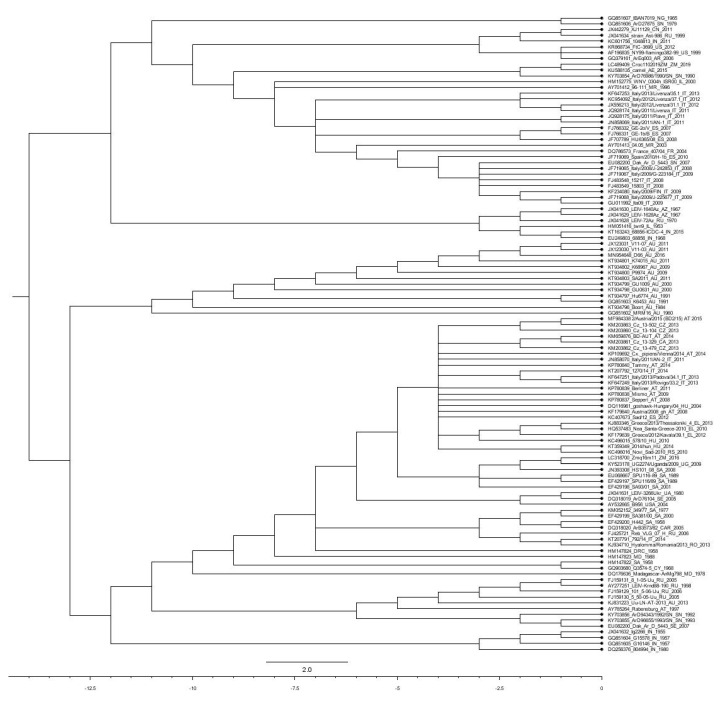
Figure 3.
Maximum-likelihood phylogenetic tree of estimating the relationships of selected West Nile virus isolates. The tree was constructed with MEGA-X software version 10.1.8. The optimal tree was obtained using Maximum likelihood method, Nearest Neighbour Interchange (NNI) inference method. The phylogeny was tested with bootstrap replicates method (N = 1000). The evolutionally distances were calculated with the general time reversible (GTR) model, uniform rates. The tree was edited with FigTree v1.4.3 software (http://tree.bio.ed.ac.uk/software/figtree/). The scale represents at the bottom represent divergence time in millions of years ago (MYA). Each sequence used was labelled by GenBank accession number_isolate/strain name_country of isolation_year of isolation.