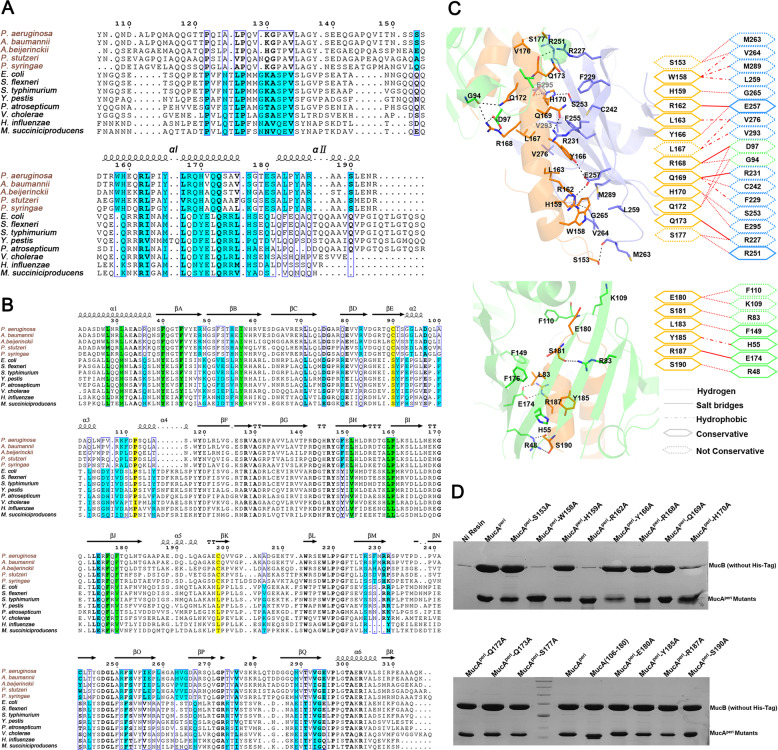
Fig. 3. Structural-based sequence alignment and specific interactions in MucAperi-MucB complex.
a Multi-sequence alignment (generated by Espript 3.0, http://espript.ibcp.fr/ESPript/ESPript/) of MucA/RseA homologous. Residues involved in protein–protein interactions are shaded in cyan. b Multi-sequence alignment of MucB/RseB homologous. Conserved hydrophobic residues that form lipid-binding pockets are shown in green, positions corresponding to residues P112, C90, and C198 in MucB (disulfide bond) are shaded in yellow. c Structures and schematics of the binding interfaces between MucAperi (in orange) αІ (top)/αII (bottom) and MucB (NTD, CTD are colored green and blue). d Pull-down assay. Incubate 20 µg MucAperi or its variants (with His-tag) with excessive MucB (without His-tag), the molar ratio of MucAperi to MucB was 1:3 to ensure excess of MucB interact with MucAperi or mutants. Fractions were eluted with solution buffer containing 300 mM imidazole and determined by 15% SDS-PAGE gel and visualized by Coomassie Brilliant Blue stain.