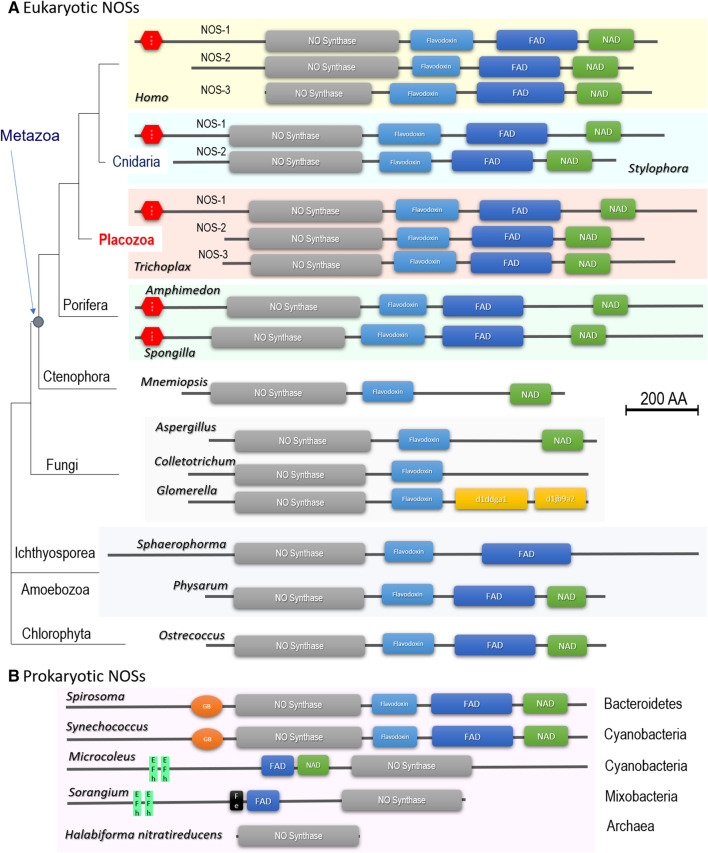
Figure 2.
Conservative domain organization of eukaryotic (A) and prokaryotic (B) NOS. All NOS are presented on the same scale, including the sizes of all domains and proteins. Grey box—NOS oxygenase domain (Pfam NO_Synthase PF02898); light blue box—Flavodoxin_1 (Pfam PF00258); dark blue box—FAD_binding_1 domain (Pfam PF00667); green box—NAD_binding_1 (Pfam PF00175). (A) Eukaryotic NOSs include representatives of 5 metazoan phyla and 4 non-metazoan lineages with their respective phylogenetic relationships and species names (see also Fig. 1 for details). Three human (Homo) NOSs with PDZ domain (red hexagon) containing neuronal NOS-1 are at the top of the figure. Trichoplax is the only know invertebrates with three NOS genes, also including PDZ domain-containing NOS-1. The ctenophore Mnemiopsis and all thee fungal NOSs apparently lost the FAD domain. Colletotrichum, Glomerella, and Sphaerophorma lost NAD domain. Interestingly, NOS in Glomerella contained unusual domains absent in other NOSs. (B) Two prokaryotic NOSs (Spirosoma and Synechoccus) have the canonical 4 domain organization, but with specific heme-binding globin domain (Pfam PF00042). The cyanobacteria Microcoleus and myxobacteria Sorangium apparently have a reverse order of canonical domains and two distinct calcium-binding motifs (EF-hand, SMART SM000054). Sorangium NOS also has BFD-like [2Fe-2S] iron-binding domain (Pfam PF04324). The Archaea representative (Halabiforma) has an NOS oxygenase domain only. The references for each particular gene and/or their sequences with relevant GeneBank accession numbers are summarized in the supplementary dataset (Excel Table).