Fig. 2.
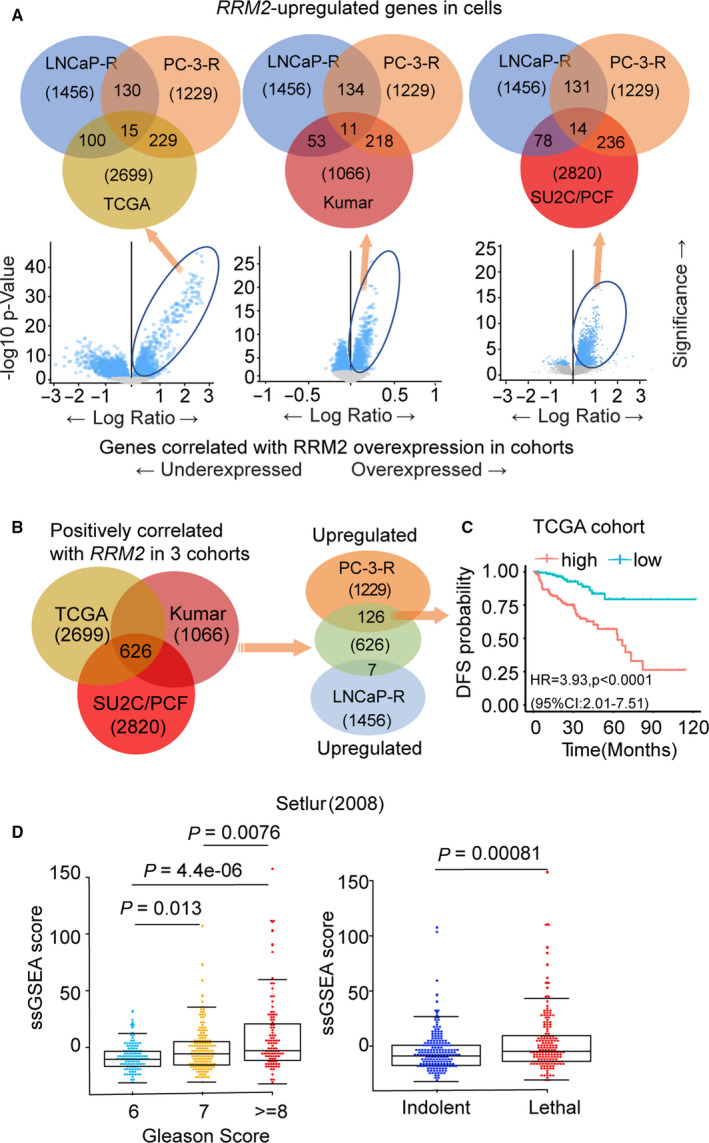
Integration of prostate cancer cell line transcriptomic data with clinical outcomes. (A) Venn diagrams (top) depicting the overlap of genes with expression that positively correlated with RRM2 levels in TCGA (left), Kumar (middle), and SU2C/PCF (right) cohorts with upregulated genes in LNCaP‐RRM2 or PC‐3‐RRM2 cells. Below, plots show the genes with expression that correlates with RRM2 expression level in each prostate cancer cohort. (B) RRM2 signature: The 626 genes with expression that correlated with RRM2 levels in the three clinical cohorts (left) were compared with genes upregulated in PC‐3‐RRM2 or LNCaP‐RRM2 (right) to identify RRM2 signature (126 genes). (C) Clinical significance of expression of the 126‐gene RRM2 signature in the TCGA cohort. Samples were ranked based on expression of the 126‐gene RRM2 signature, and Kaplan–Meier curves were used to estimate survival differences between patients in the top and bottom 25th percentiles of expression. The log‐rank test was calculated to determine significance. Cox proportional hazard regression was performed, adjusting for clinical and demographic factors. (D) Association between RRM2 signature (126 genes) level with Gleason score (left) and lethality (right) in the Setlur cohort. Comparisons between groups were performed using Wilcoxon's rank‐sum test.
