Figure 4.
Identification of SMXL Domains Mediating Nuclear Localization and KAR1-Induced Degradation.
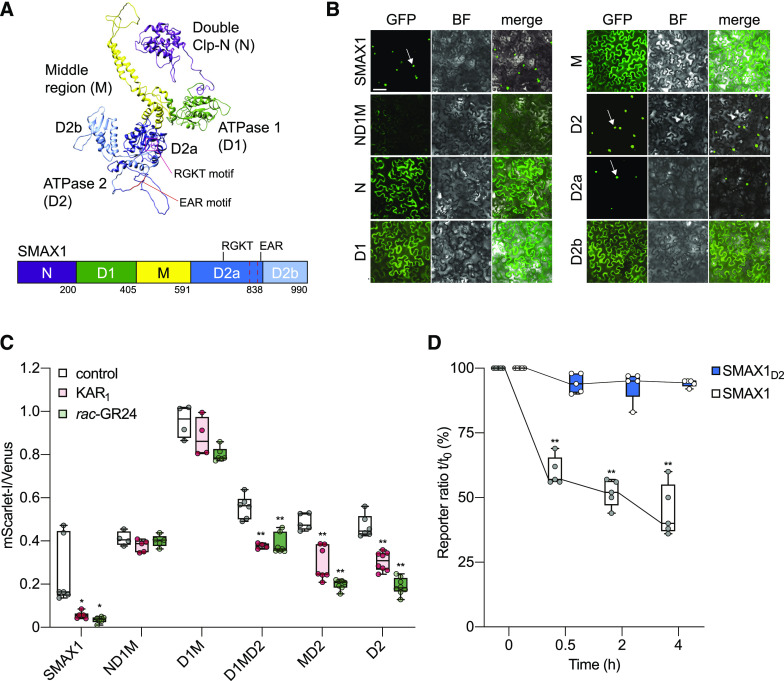
(A) Model of the three-dimensional structure of the SMAX1 protein predicted by Phyre2. Major SMAX1 domains are indicated: (N (purple), D1 (green), M (yellow), and D2 (blue). D2 is divided into D2a (dark blue) and D2b (light blue) subdomains. RGKT motif and EAR motif are highlighted in red. The red dashed box indicates a putative NLS sequence at amino acids 797 to 824.
(B) Subcellular localization of SMAX1 domains fused to GFP after transient expression in N. benthamiana epidermal cells. Expression was driven by the 35S promoter. Bar = 50 μm. BF, bright field.
(C) Relative fluorescence of NLS-SMAX1 domains in the pRATIO1212 system after transient expression in N. benthamiana and overnight treatment with 10 μM KAR1, 10 μM rac-GR24, or 0.02% (v/v) acetone control. n = 5 to 8 leaf discs. *P < 0.01, **P < 0.001, Mann–Whitney U test, comparisons to control treatment.
(D) Time course of SMAX1 and SMAX1D2 stability in N. benthamiana after addition of cycloheximide (150 µg/mL). SMAX1 and SMAX1D2 abundance was monitored through the pRATIO1212 system. The reporter:reference ratio at each time point was scaled to the ratio measured at time 0 to show relative abundance over time. n = 5 leaf discs. **P < 0.001, paired t test, comparison to time 0.