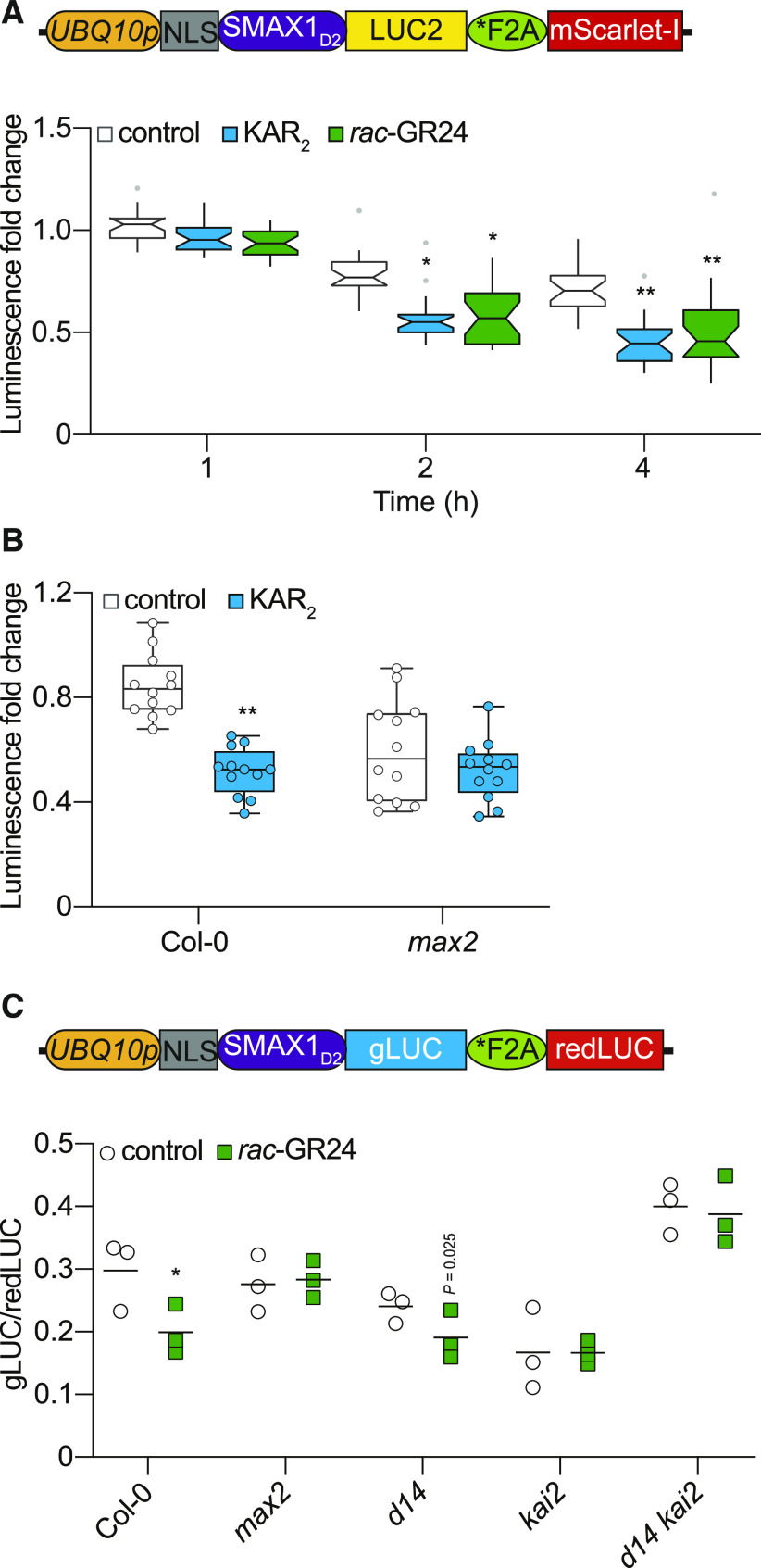
Figure 5.
MAX2- and KAI2-Dependent Degradation of SMAX1D2 in Arabidopsis.
(A) Diagram of the pRATIO2251-SMAX1D2 for ratiometric detection of SMAX1D2 in Arabidopsis transgenic lines. The UBQ10 promoter drives expression of a multicistronic transcript encoding an NLS-SMAX1D2-LUC2 fusion that is separately translated from the mScarlet-I fluorescent protein due to the *F2A peptide. Bioluminescence of SMAX1D2-LUC2 in homozygous Arabidopsis transgenic lines treated with 10 μM KAR2, 10 μM rac-GR24, or 0.02% (v/v) acetone control. Fold changes in bioluminescence relative to time 0 (immediately after treatment) for each seedling are shown. Boxplot notches approximate the 95% CI for the median. n = 24. *P < 0.01, **P < 0.001, two-way analysis of variance with Bonferroni’s test, comparisons to control treatment.
(B) Bioluminescence of SMAX1D2-LUC2 in the Col-0 and max2 backgrounds. Seedlings were treated with 10 µM KAR2 or acetone control for 4 h. Fold changes in bioluminescence relative to time 0 are shown. n = 12. **P < 0.001, Mann–Whitney U test, comparisons to control treatment.
(C) Diagram of the pMD19-SMAX1D2 for ratiometric detection of SMAX1D2 in Arabidopsis protoplasts. In pMD19-SMAX1D2, which is a nonbinary plasmid, the UBQ10 promoter controls expression of a multicistronic transcript encoding an NLS-SMAX1D2-gLUC fusion that is separately translated from a red-shifted firefly LUC protein due to a *F2A peptide. SMAX1D2 abundance in protoplasts isolated from Col-0, max2, d14, kai2, and d14 kai2, transformed with a dual-LUC ratiometric reporter (pMD19-SMAX1D2), recovered for 24 h, and treated with 10 μM rac-GR24 or acetone control for 4 h. SMAX1D2 was fused to gLUC and redLUC bioluminescence was used as a reference. gLUC:redLUC ratios of three independent protoplast preparations and transformations are shown; bar indicates mean. Results of a representative experiment are shown. *P < 0.01, unpaired t test, comparisons to control treatment.