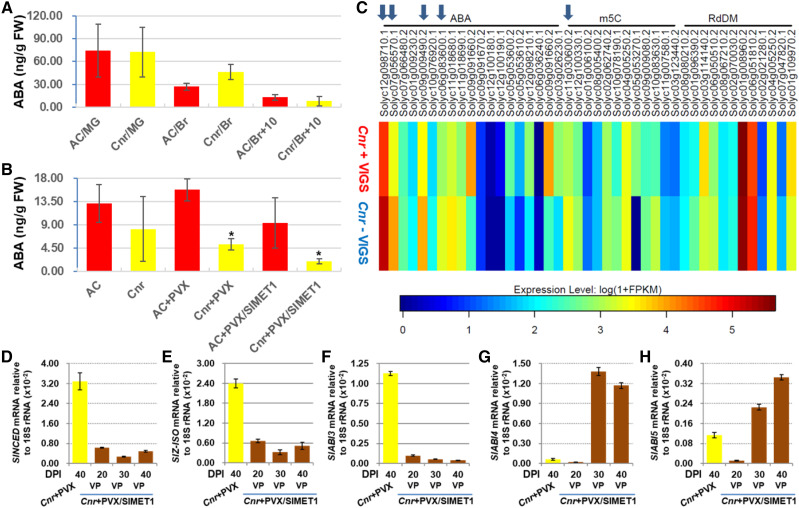
Figure 2.
Influences of SlMET1 silencing on ABA biosynthesis and ABA-related gene expression. A, ABA at different fruit developmental stages. Detection was carried out on four biological fruit samples collected at mature green (30 DPA), breaker (36 DPA on average), and 10 d after breaker (47 DPA on average) stages. Data are shown as means ± sd (n = 4). Red bars represent AC and yellow bars represent Cnr. FW, Fresh weight. B, SlMET1 knocked down by VIGS affects ABA accumulation. ABA assays were on four different AC or Cnr fruits mock inoculated or injected with PVX or PVX/SlMET1 at 30 DPI (equivalent to 47 DPA on average). Significant reduction of ABA levels was seen in viviparous seeds/seedlings from SlMET1-KD (Cnr+PVX/SlMET1) Cnr fruits when compared with Cnr fruits injected with PVX (Cnr+PVX; one-way ANOVA, P < 0.0016, as indicated by asterisks). Data are shown as means ± sd (n = 4). C, Identification of epigenetically regulated differentially expressed genes (epiDEGs) by comparative RNA-seq and comparative WGBS. Arrows from left to right indicate four epiDEGs (SlZ-ISO, SlNCED, SlABI5, and SlABI3) in addition to SlMET1, which was knocked down by VIGS. The four epiDEGs along with SlABI4 were chosen for further investigation due to their pivotal role in ABA biosynthesis and response. Names and SOL identifiers for these epiDEGs are detailed in Supplemental Table S2. D to H, Effects of SlMET1-KD on the expression of ABA biosynthesis and ABA-responsive genes. The expression of SlNCED (D), SlZ-ISO (E), and SlABI3 (F) was significantly down-regulated (one-way ANOVA, P < 0.05) at 20, 30, or 40 DPI, while that of SlABI4 (G) and SlABI5 (H) was significantly up-regulated (one-way ANOVA, P < 0.05) at 30 or 40 DPI in viviparous seeds/seedlings from PVX/SlMET1-injected Cnr fruits, compared with mature seeds from PVX-injected control Cnr fruits. RT-qPCR assays were performed on three different samples collected at different DPI as indicated. RNA transcript level was normalized against 18S rRNA. Data are shown as means ± sd (n = 3).