Figure 1.
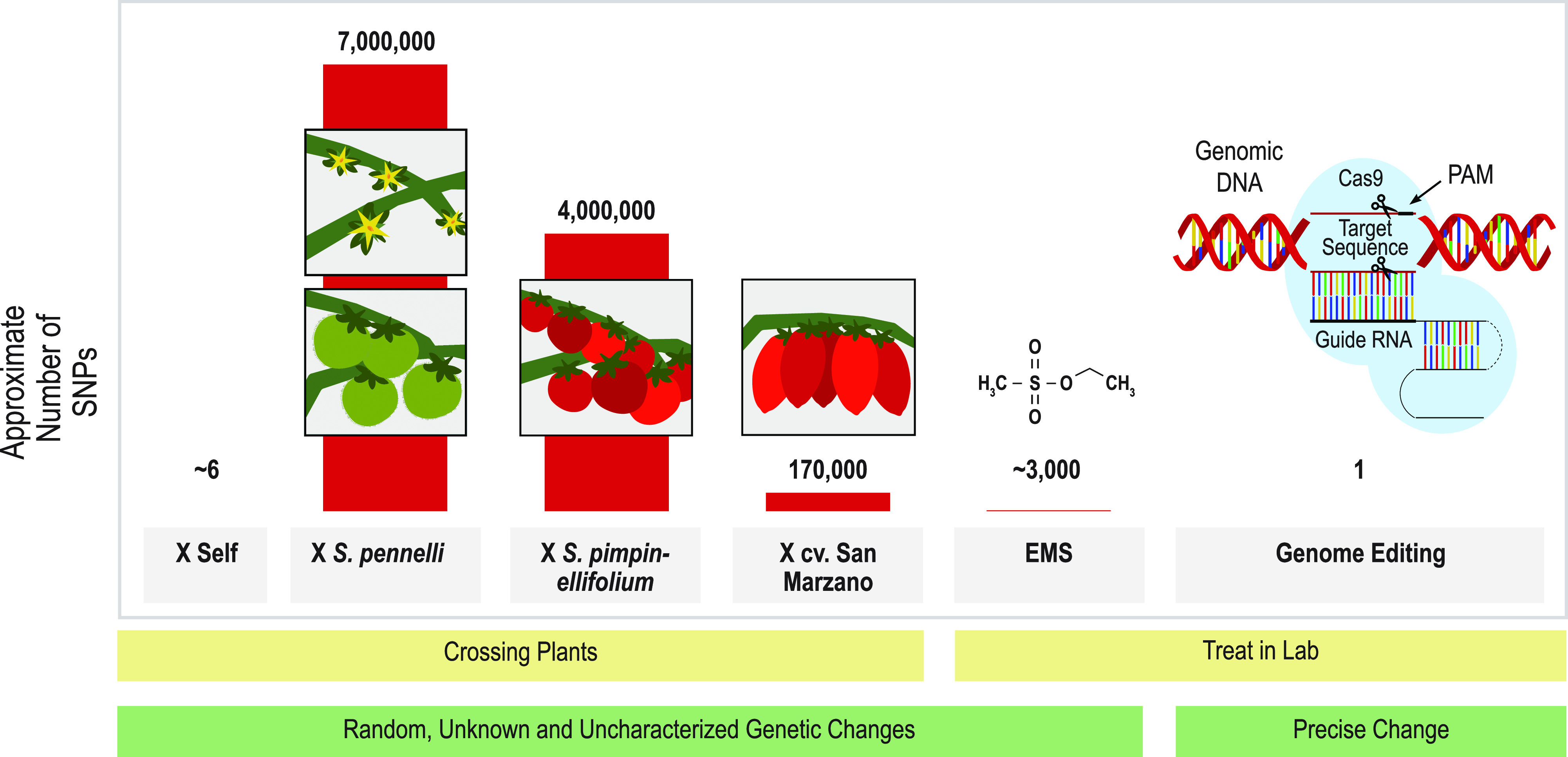
Comparison of the average number of SNPs and indels per genome (individual) introduced into tomato by different breeding strategies. The data represent approximate numbers of SNPs between the genome sequence of S. lycopersicum (Heinz 1706 reference genome) and other cultivars or wild relatives which have been used in breeding of modern tomato cultivars. After selfing (× Self), each individual plant in the next generation will have (on average) approximately six random SNPs (assuming the de novo rate of spontaneous heritable SNP formation in tomato is similar to that of lab-grown Arabidopsis (Ossowski et al., 2010). Most modern elite tomato lines commercially grown typically have four or more disease-resistance genes that have been introgressed by crossing with wild tomato species such as Solanum pennellii or Solanum pimpinellifolium (Foolad, 2007). These initial elite × wild species hybridization events introduced millions to tens of millions of SNPs in addition to indels, CNVs, and PAVs (Aflitos et al., 2014). Crosses with more closely related domestic tomato lines or landraces (i.e. cv San Marzano) will introduce (on average) hundreds of thousands of SNPs/indels (Ercolano et al., 2014). Creating random variation by treating seeds with the chemical mutagen ethyl methanesulphonate (EMS) typically introduces thousands of SNPs per individual (Minoia et al., 2010). Treating cells with a well-designed gene editing reagent (including a gRNA homologous to target sequence and adjacent PAM) can create a single SNP or indel at a precise, predetermined location. Crossing with wild or closely related species can also introduce additional indels, CNVs, and PAVs into the genome, which is not considered in this figure.
