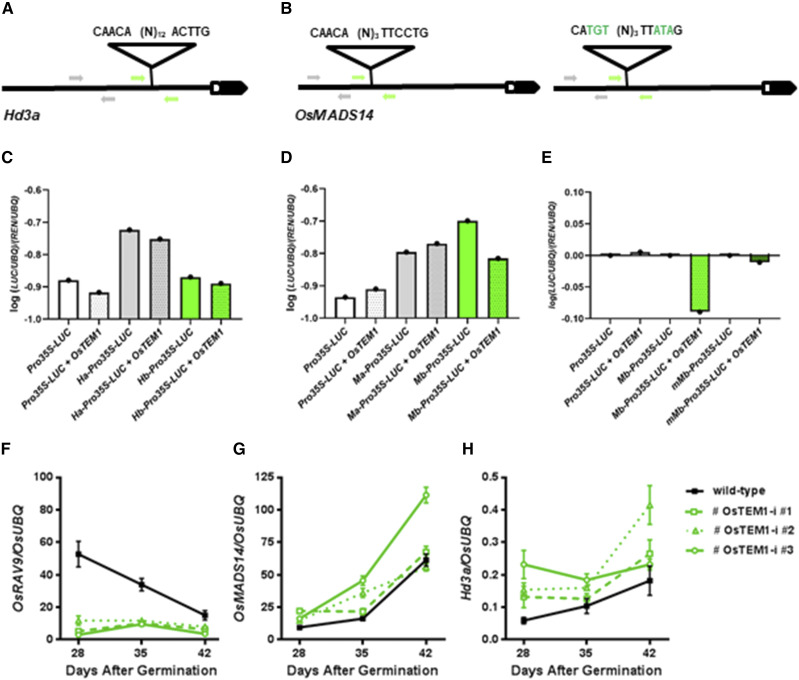
Figure 5.
Interaction between OsRAV9/OsTEM1 and floral activators. A, Noncanonical RAV binding site in the promoter of Hd3a, 785 bp upstream of the transcription starting site. B, Perfect RAV binding site in the promoter of OsMADS14, 2250 bp upstream of the transcription starting site, and mutated version (right). Arrows represent oligonucleotides used to amplify fragments of ProHd3a (H) and ProOsMADS14 (M) without RAV binding sites (Ha and Ma, in gray) and with RAV binding sites (Hb and Mb, in green). C to E, Transactivation activity of OsTEM1 in transiently transformed protoplasts, reported as ratio of transcript levels of LUC and REN reporter genes relative to UBQ. C, RT-qPCR of protoplasts cotransformed with Pro-35S:OsTEM1 and reporter vectors containing sequences of ProHd3a (Ha in gray, Hb in green). D, RT-qPCR of protoplasts cotransformed with Pro-35S:OsTEM1 and reporter vectors containing sequences of ProOsMADS14 (Ma in gray, Mb in green). E, Analysis of protoplasts cotransformed with Pro-35S-OsTEM1 and reporter vectors containing sequences of ProOsMADS14 (Mb in green, mutated Mb in dark green). Values are the mean of three independent replicates. F to H, Expression analysis of genes involved in heading date in independent T2 lines (green) compared with wild-type plants grown for 28, 35, and 42 d under inductive conditions. F, Down-regulation of OsRAV9/OsTEM1 in silencing lines (green). G and H, Up-regulation of the floral activators OsMADS14 and Hd3a in silencing lines (green lines) compared with wild-type (black line). Expression data are mean value of three biological replicates with three technical replicates each, and error bars represent sd.