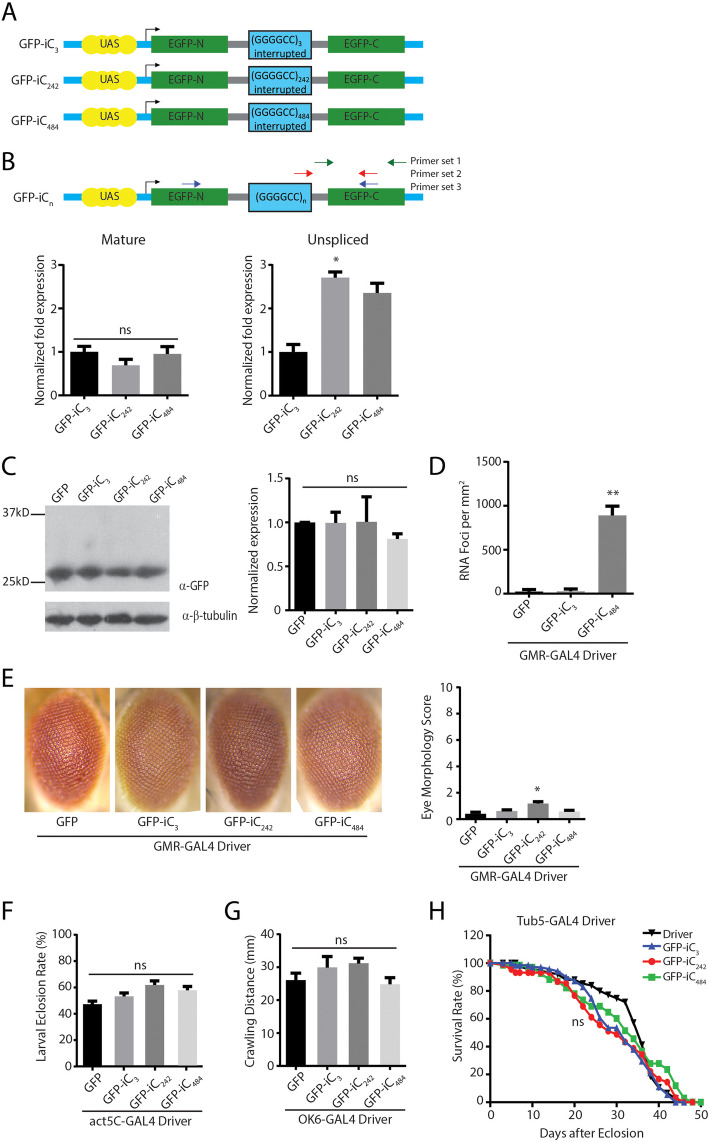
Fig. 1.
Intronic G4C2 repeats influence splicing and elicit RNA foci in Drosophila.a Schematic of constructs used to generate transgenic fly lines. The Prospero fly intron 1 (gray lines) was introduced into UAS-GFP (blue lines). Either three G4C2 repeats or serial (G4C2)21–28 repeat units separated by 14 nt interruptions were serially inserted into the intron. b Locations of the primer pairs used for measuring unspliced, spliced, and total mRNA are show in the schematic. Quantification of the expression of the mature (left) and unspliced (right) GFP product in the indicated fly lines (right), n = 9. c Western blot of lysates from heads of G4C2 intronic repeat flies (left) with quantification of GFP normalized to beta-tubulin (right), n = 5. d Quantification of RNA foci in the retina of the indicated intronic fly, n = 10. e Representative eye phenotypes from flies of the indicated genotypes crossed to GMR-GAL4 to drive expression in developing ommatidia (left) and quantification of eye phenotype scores (right). f Quantification of the number of progeny carrying the transgene after flies of the indicated genotypes were crossed to a ubiquitous driver (act5c-GAL4). g Quantification of the distance crawled by 3rd instar larvae from the crosses of the indicated fly genotypes to a larval motor neuron specific driver (OK6-GAL4). h Flies carrying the indicated transgenes were crossed to a Tubulin Geneswitch driver (Tub5) to allow ubiquitous expression post eclosion. Adult male flies were placed on food containing RU-486 to activate gene expression and their viability was tracked over 50 days. Graphs represent means ± SEM. * p < 0.05; ** p < 0.01 by Kruskal-Wallis after Dunn’s correction for multiple comparisons. The number of flies for each genotype for E, F, G and H was > 100