Figure 3. GiGA1 Activates GIRK1/GIRK2 Channels through an Alcohol Pocket in a G-Protein-Independent Manner.
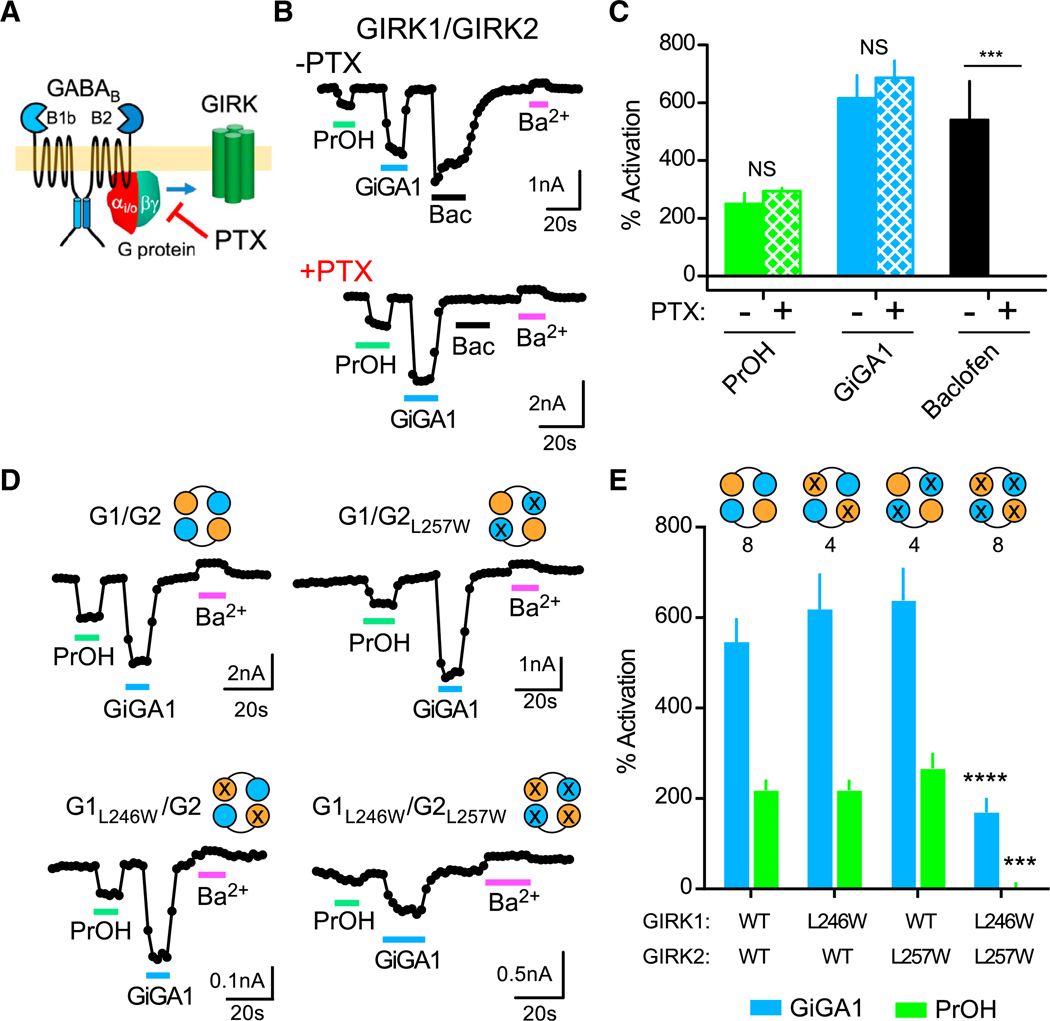
(A) Cartoon shows activation of the GIRK channel via the G-protein-dependent pathway, which is blocked by PTX.
(B) Traces show the current response to PrOH (100 mM), GiGA1 (100 μM), baclofen (100 μM), and Ba2+ (1 mM) in HEK cells transfected with cDNA for GABAB1b/B2 receptors and GIRK1/GIRK2c tandem dimer with or without the PTX S1 subunit.
(C) Bar graph shows the mean change in percentage of activation for GIRK1/GIRK2 channels with PrOH (green), GiGA1 (blue), or baclofen (black) in the absence or presence of the PTX S1 subunit. ***p = 0.0006; F(2, 5) = 25.52 (baclofen × PTX); mixed-effects model (restricted maximum likelihood [REML]) with Bonferroni post hoc (n = 4 (−PTX), 5 (+PTX)).
(D) Traces for the WT GIRK1/GIRK2 channel and the indicated alcohol pocket mutation (GIRK1L246W/GIRK2L257W) (Aryal et al., 2009) in response to PrOH (100 mM), GiGA1 (100 μM), and Ba2+ (1 mM). The schematic indicates the subunit that has a mutation; the orange circle is GIRK1, and the cyan circle is GIRK2.
(E) Bar graph shows the mean percentage of activation response to GiGA1 and PrOH. ***p = 0.0004, ****p < 0.0001 versus GIRK1/GIRK2 WT; two-way repeated-measures ANOVA with Dunnett’s multiple comparison test (F(3, 20) = 5.298; ***p = 0.0075 activator × mutation). n is indicated on the graph.
Error bars represent SEM on the graphs.
