Figure 4. GiGA1 Activates GIRK1/GIRK2 Channels through a Different Mechanism from ML297.
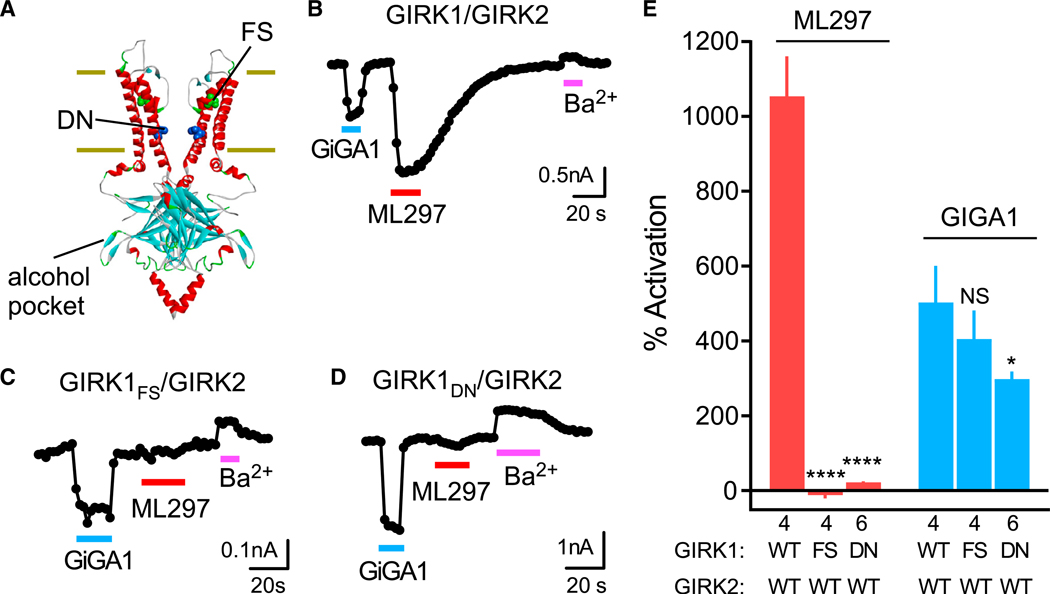
(A) Structural model of GIRK2 (PDB: 4KFM) shows the alcohol pocket and location of homologous amino acids in GIRK1 implicated previously in ML297 activation (FS (green), GIRK1F137 to ~GIRK2S148; DN (blue), GIRK1D173 to ~GIRK2N184).
(B–D) Traces show the current response of the WT GIRK1/GIRK2 (B), GIRK1FS/GIRK2 (C), and GIRK1DN/GIRK2 (D) channels to GiGA1 (100 μM), ML297 (10 μM), and Ba2+ (1 mM). Both mutants abolished the ML297 response, but not the activation with GiGA1.
(E) Bar graph shows the mean percentage of activation with GiGA1 and ML297. ****p < 0.0001, *p = 0.0374 versus GIRK1/GIRK2 WT; two-way repeated-measures ANOVA with Dunnett’s multiple comparison test (F(2, 11) = 106.3; p < 0.0001; activator × mutation). n is indicated on the graph. Error bars represent SEM on the graph.
