Figure 4.
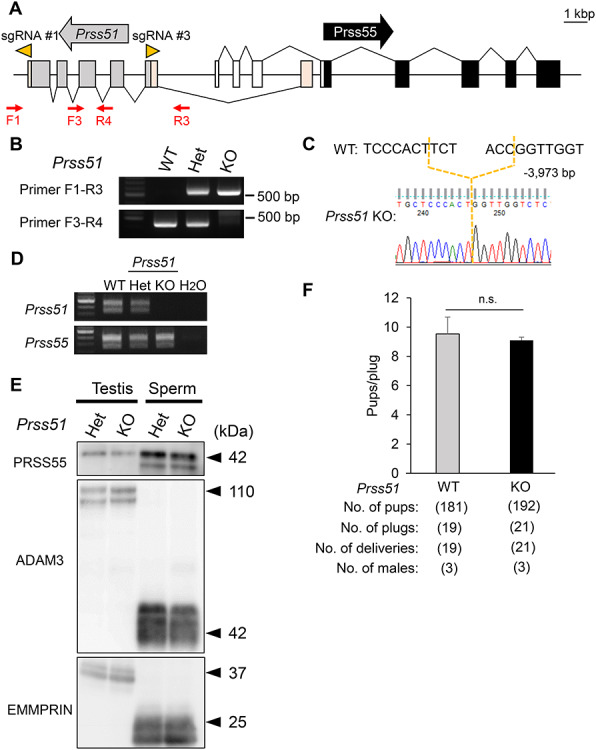
Male mice lacking Prss51 are fertile. A. Genomic structure and KO strategy to create mouse Prss51 KO mice. The direction of the arrows, in gray (Prss51) and black (Prss55), indicates the direction of transcription. The direction of the arrowheads (orange) indicates the direction of sgRNA (#1 and #3). Gray boxes, Prss51 coding region; light orange boxes, Prss51 noncoding region; black boxes, Prss55 coding region; white boxes, Prss55 noncoding region. F1 and F3, forward primers for genotyping; R3 and R4, reverse primers for genotyping. Scale bar, 1 kbp. B. Genotyping with genomic PCR. Primers shown in panel A were used for the PCR. WT, wild type; Het, heterozygous; KO, Prss51 homozygous KO. C. DNA sequencing. The sequence of the PCR amplicon was analyzed. A region of 3973 bp was deleted in Prss51 KO mice. D. Expression of mouse Prss51 and Prss55 was examined by RT-PCR using RNA isolated from testes. WT, wild type; Het, heterozygous; KO, Prss51 homozygous KO. E. Detection of PRSS55 and ADAM3 by immunoblotting. EMMPRIN was used as a control. Testis, whole testis; sperm, cauda epididymal spermatozoa; Het, heterozygous; KO, Prss51 homozygous KO. Black arrowheads, molecular weights (kDa). F. Fertility is presented as pups/plug: the number of total pups born divided by the number of vaginal plugs in females after natural mating. (9.5 ± 1.2 for WT females mated with WT males and 9.1 ± 0.2 with Prss51 KO males). N.s., no significance (P > 0.05). Error bars, mean ± SD.
