Figure 2. Summary of the model organism ortholog table and multi-species alignment for TBX2.
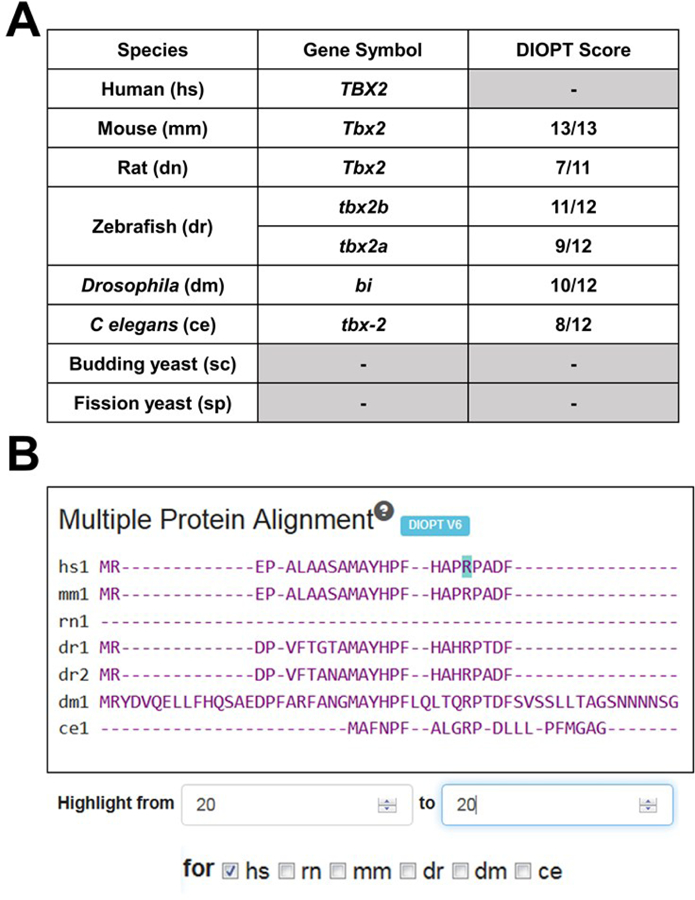
A) MARRVEL selects the top ortholog candidate for each species based on the DIOPT tool. For example, a DIOPT score of 10/12 shown for the Drosophila bi gene means 10 out of 12 orthology prediction programs used by DIOPT predicted that bi is the most likely fly ortholog of human TBX2. Since 25% of genes are duplicated in zebrafish compared to human, MARRVEL displays two paralogous genes (in this case tbx2a and tbx2b) when this is applicable. B) Snapshot of the multi-species alignment window. By selecting a specific organism [in this case human (hs)] and entering the amino acid of interest, one can highlight the specific amino acid in teal. In this example, p.R20 of human TBX2 seems to be conserved in mouse (mm1), both zebrafish orthologs (dr1 and dr2), Drosophila (dm1) and C. elegans (ce1). Rat Tbx2 does not seem to align well compared to other species, most likely due to the isoform used by the DIOPT to perform the multi-species alignment.
