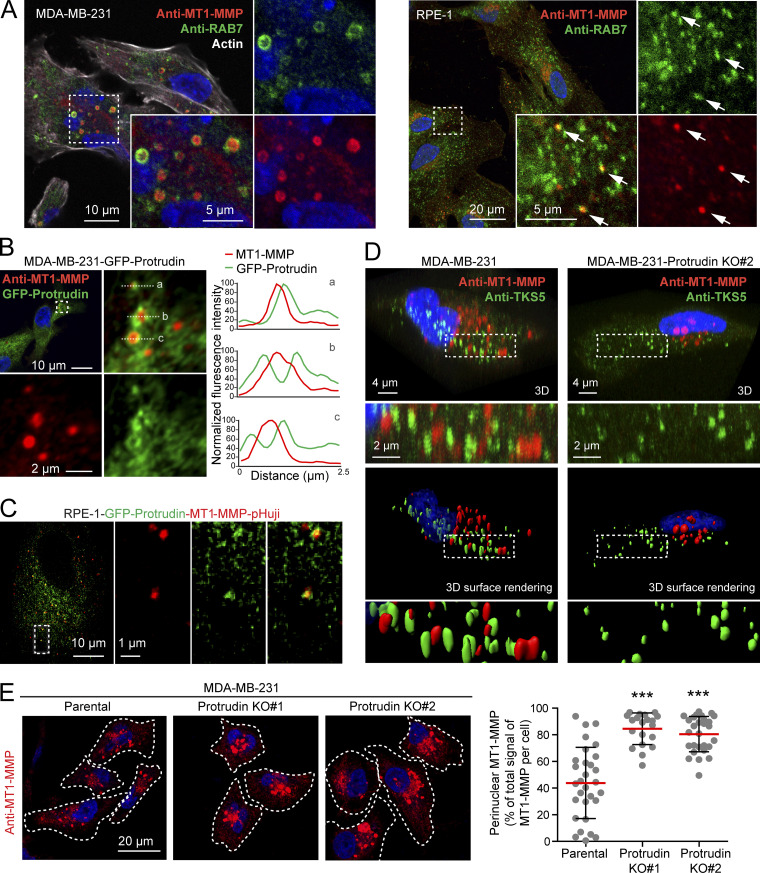
Figure 5.
Protrudin forms contact sites with MT1-MMP–positive LE/Lys and regulates their intracellular localization. (A) Confocal micrographs of MDA-MB-231 and RPE-1 cells showing that endogenous MT1-MMP localizes to RAB7-positive endosomes. Arrows show colocalization of MT1-MMP and RAB7 in RPE-1 cells. Manders overlap coefficient shows that 56% (±12 SD) of MT1-MMP positive pixels overlap with Rab7-positive pixels in MDA-MB-321 cells (n = 15 cells), and 33% (±8 SD) in RPE-1 cells (n = 13 cells). (B) Confocal micrographs of MDA-MB-231-GFP-Protrudin cells showing contact sites between GFP-Protrudin and endogenous MT1-MMP. Graphs show three examples of contact sites between GFP-Protrudin in the ER and MT1-MMP–positive endosomes visualized by fluorescence intensity profiles. (C) Still image from Video 1 of RPE-1-GFP-Protrudin-MT1-MMP-pHuji cells showing that GFP-Protrudin forms contact sites with MT1-MMP-pHuji–positive vesicles. (D) MDA-MB-231 and MDA-MB-231-Protrudin KO#2 cells were grown on coverslips, immunostained for MT1-MMP and TKS5, and analyzed by confocal microscopy. Z-stacks and Imaris surface 3D renderings show the subcellular localization of MT1-MMP in relation to TKS5-positive invadopodia. Note the perinuclear clustering of MT1-MMP and the undersized TKS5-positive invadopodia in Protrudin-KO#2 cells. (E) MDA-MB-231 and MDA-MB-231-Protrudin-KO cells were grown on coverslips, immunostained for MT1-MMP, and analyzed by confocal microscopy. Images and graph show the perinuclear clustering of MT1-MMP–positive endosomes in Protrudin-KO cells. Each plotted point represents one cell. Values represent mean ± SD. Number of cells analyzed: parental, n = 30; KO#1, n = 20; KO#2, n = 28 from three independent experiments. ***, P < 0.001, Kruskal-Wallis, Dunn’s post hoc test. Note that the big variation in MT1-MMP localization seen between parental cells is diminished in Protrudin-KO cells where MT1-MMP vesicles mainly cluster perinuclearly.